Discovery of sphingosine 1-O-methyltransferase in rat kidney and liver homogenates1
Introduction
Sphingolipids are ubiquitous constituents of eukaryotic membranes. Their metabolism is a highly dynamic process that generates various secondary messengers, including ceramide, sphingosine, and sphingosine 1-phosphate (S1P)[1–3]. Ceramide is deacylated by ceramidases to generate sphingosines, which can be phosphorylated by sphingosine kinases to produce S1P[1]. It is now generally understood that ceramide/sphingosine and S1P have contrasting effects on the stress response[1–4]. Sphingolipids, including sphingosine, S1P, and ceramide, have been studied intensively over the last 2 decade[1,5]. N,N-dimethyl-D-erythro-sphingosine (DMS), a methylated derivative of sphingosine, is found naturally in cells, and the enzymes that convert sphingosine to DMS have been detected in several tissues[6–8]. DMS was first reported as a protein kinase C inhibitor along with D-erythro sphingosine[9–11]. Later, Spiegel et al reported its specific inhibitory action on the sphingosine kinase and excluded its inhibitory action on protein kinase C[12,13]. Thereafter, DMS has been used as a specific inhibitor of the sphingosine kinase. Its action on tumor cell migration and cancer cell growth have also been reported and are a fundamental basis for the development of chemotherapeutic agents[14–16]. However, sphingosine N-methyltransferase, the enzyme responsible for DMS production, has not been well characterized.
On a preliminary note, the enzymatic methylation of sphingosine in the mouse brain homogenate with S-adenosyl-L-methionine (SAM) as the methyl donor was reported[6]. The N-methyltransferase responsible for DMS synthesis was shown to be inde-pendent of the N-methyltransferase that converts phosphatidylethanolamine to phosphatidylmonomethyl-ethanolamine, phosphatidyldimethyl-ethanolamine, and phosphatidylcholine. The former was only detectable in the brain (not in the liver), while the latter was present in both the brain and liver, and probably in all cells[17,18]. In this study, we tried to identify this enzymatic activity; however, we found another methyltransferase activity for sphingosine in the rat kidney and liver homogenates that transfers a methyl group from SAM to the C1 OH group of sphingosine.
Materials and methods
Materials Cold SAM was purchased from Sigma (St Louis, MO, USA). D-erythro-sphingosine (synthetic), DMS, N-methyl-D-erythro-sphingosine (MMS), and N,N,N-trimethyl-D-erythro-sphingosine (TMS) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). S-adenosyl-L-(methyl-3H) methionine was procured from Amersham (GE Healthcare UK, Buckinghamshire, UK), while E3 enhancer was purchased from PerkinElmer Life & Analytical Sciences (Boston, MA, USA). Silica gel high performance thin layer chromatography (HPTLC) (60 F254, 20×20) was obtained from Merck (Darmstadt, Germany), and Kodak Medical X-ray film was purchased from Eastman Kodak (Rochester, NY, USA). D-threo-sphingosine, L-threo-sphingosine, and L-erythro-sphingosine were from Matreya (Pleasant Gap, PA, USA). C3-deoxy-sphinganine was kindly provided by SK CHUNG at Postech (Pohang, Gyungbuk, Korea)[19]. C1-deoxy-sphinganine was synthesized by WK LEE at Sogang University (Seoul Korea). All other materials were purchased from Sigma–Aldrich Korea (St Louis, MO, USA).
Preparation of tissue homogenates Rat tissue homogenates were prepared as described[6] with some modifications. Spleen, lung, thymus, heart, brain, and liver tissues were removed from Sprague-Dawley rats (n=2−3) and separately washed in cold phosphate-buffered saline. The tissues were placed in 10 mL cold 20 mmol/L Tris-HCl (pH 7.5) and 2 mmol/L EDTA solution, and homogenized with a Dounce homogenizer at 4 °C. The homogenate was centrifuged at 500×g for 10 min to remove unbroken cell debris, and the supernatant was used as an enzyme source for sphingosine methyltransferase.
Assay method for sphingosine methyltransferase We employed the same assay conditions as Igarashi and Hakomori to observe the enzymatic activity of sphingosine methyltransferase[6]. The complete reaction mixture included 40 mmol/L Tris-HCl (pH 7.8), 3 mmol/L MgCl2, 10 µmol/L 3H-SAM (1 mCi/mmol), 300 µmol/L sphingosine, and tissue homogenates (~5 mg of protein) in a final volume of 1 mL. Protein amounts were determined by the Micro BCA protein assay reagent kit (Pierce, Rockford, IL, USA) with bovine serum albumin as a standard. Sphingosine was added to the reaction mixture as an exogenous substrate (300 µmol/L) and incubated for 1 h at 25 °C. The reaction was started by the addition of the tissue homogenate (~5 mg of protein) to the preheated reaction solution. After 1 h, the incubation was terminated by adding 100 µL of 10 mol/L HCl. To extract the metabolites, 5 mL chloroform:methanol mixture (2:1) was added and the tubes were capped and shaken.
The organic extracts of the reaction mixture were washed twice with the upper phase. The organic phase from each tube was then transferred and dried under nitrogen gas. The samples were reconstituted in 50 µL of chloroform:methanol mixture and spotted on 20×20 HPTLC silica gel plates for TLC. The plates were developed in chloroform:methanol:ammonium hydroxide (80:20:2) at room temperature and were allowed to air dry. Spots were visualized with an E3 enhancer spray, and autoradiography was determined with Kodak medical X-ray film (3 d exposure at –80 °C).
Results
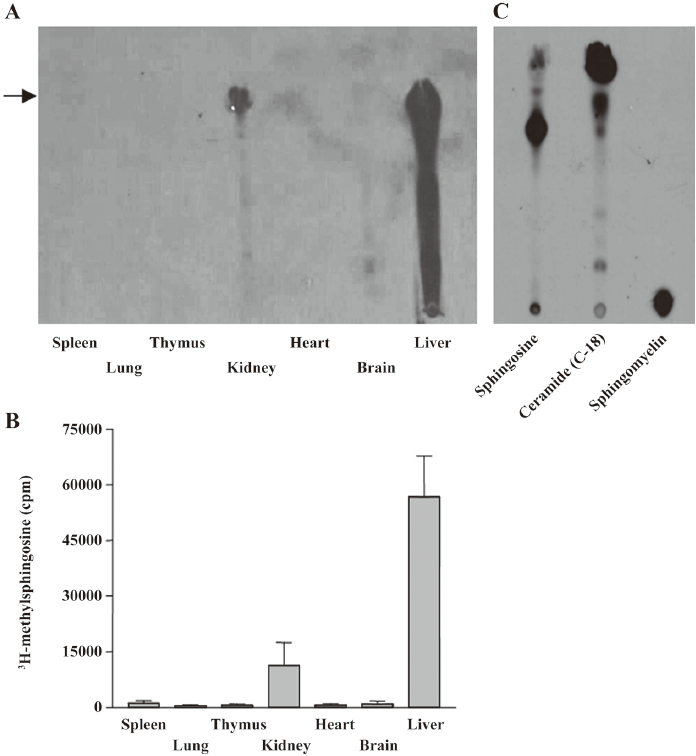
Sphingosine methyltransferase activity in kidney and liver Sphingosine methyltransferase activity was measured in rat tissue homogenates from the spleen, lung, thymus, kidney, heart, brain, and liver. As shown in Figure 1, enzymatic activity was detected in the kidney and liver homogenates. Standard sphingosine and methylated sphingosines moved to the position indicated by the arrow. In the liver homogenate, methylated lipids were also observed between methylated sphingosine and the origin of the HPTLC, implying further metabolism of the methylated sphingosine to sphingolipid metabolites, such as ceramide or sphingomyelin. In other homogenates, including the brain, enzymatic activity was undetectable, in contrast to the previous report in which sphingosine N-methyltransferase activity was detected in the mouse brain, but not in the liver[6]. The enzymatic activity in the kidney homogenate was measured under various pH conditions to find the optimum pH. The enzymatic activity was greatest at pH 7.8 (data not shown). To optimize the conditions for enzymatic activity, we measured the activity at 5, 15, 30, 60, and 120 min. The 60 min incubation was found to be the best (data not shown). Thus, subsequent experiments were conducted at pH 7.8 for 60 min.
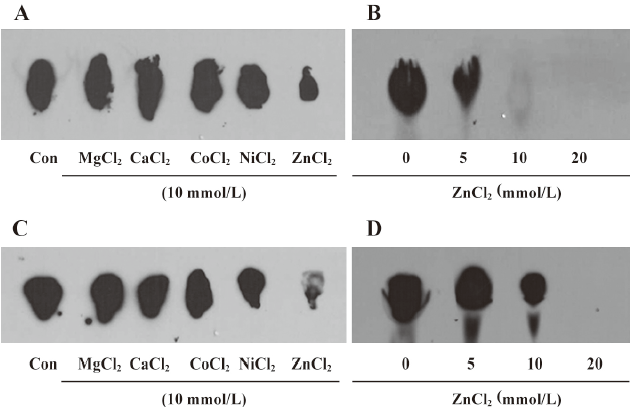
Effect of metal ions on sphingosine methyltransferase The effects of MgCl2, CaCl2, CoCl2, NiCl2, and ZnCl2 on enzymatic activity were tested. ZnCl2 strongly inhibited enzymatic activity, but other metal ions did not influence the enzymatic activity in the kidney and liver homogenates (Figure 2A and 2C). The inhibitory effect of ZnCl2 was concentration dependent (Figure 2B and 2D).
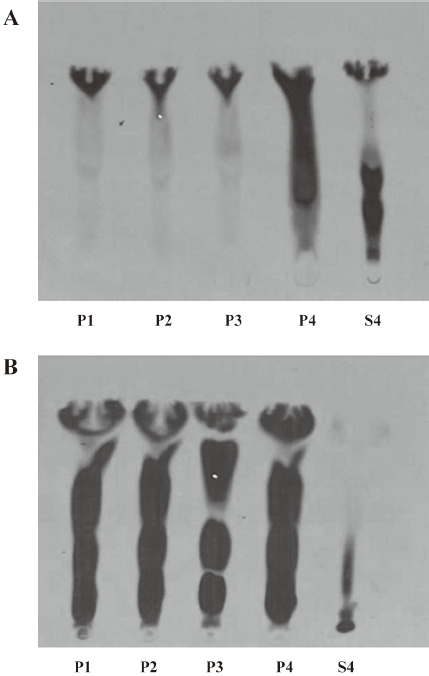
Fractionation of enzymatic activity by centrifugation The kidney and liver homogenates were fractionated by sequential centrifugation at 1000×g for 10 min, 3000×g for 10 min, 10 000×g for 20 min, and 100 000×g for 40 min. After centrifugation, we obtained 4 pellets (P1, P2, P3, and P4) and 1 supernatant (S4). Fraction P1 contained nuclei, plasma membrane sheets, heavy mitochondria, and unbroken cells. Fraction P2 generally contained heavy mitochondria and plasma membrane fragments. Fraction P3 consisted of mitochondria, lysosomes, peroxisomes, Golgi membranes, and some rough endoplasmic reticulum, while fraction P4 contained membrane vesicles (microsomes) from smooth and rough endoplasmic reticulum. Fraction S4 contained all of the soluble components from the cytoplasm.
In the kidney homogenate, the centrifugation fractions, pellets P1, P2, P3, and P4, exhibited enzymatic activity along with the supernatant (S4; Figure 3A). In the liver homogenate, fractions P1, P2, P3, and P4 showed stronger enzymatic activity than fractions from the kidney homogenate; there was no activity in the liver supernatant (S4; Figure 3B).
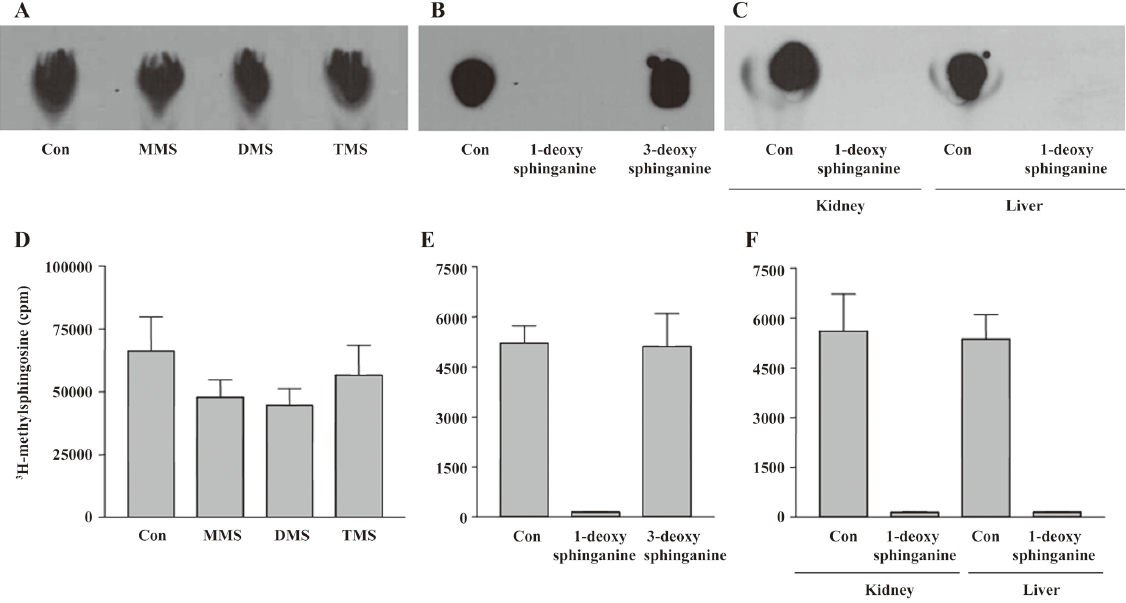
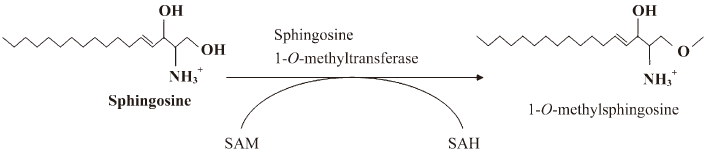
Substrate specificity Substrate specificity was next tested. MMS, DMS, and TMS were substrates for the enzyme (Figure 4A, 4D). Because TMS could not be further methylated on the C2 ammonium moiety, the enzymatic activity does not involve N-methylation on the C2 NH2 group of sphingosines. Therefore, we tested whether 1-deoxy-sphinganine and 3-deoxy-sphinganine were substrates for the enzyme. As shown in Figure 4B, 3-deoxy-sphinganine was methylated, which indicated that O-methylation on C3 OH did not occur. Finally, we synthesized 1-deoxy-sphinganine and tested it as a substrate. 1-Deoxy-sphinganine was not methylated, suggesting that methylation occurs on the C1 OH group of sphingosines (Figure 4B, 4E). As shown in Figure 4C and 4F, no methylation was observed also in the liver homogenates with 1-deoxy-sphinganine, conclusively indicating that the enzyme is sphingosine 1-O-methyltransferase (Figure 5).
Discussion
Initially, we attempted to detect sphingosine N-methyltransferase activity by converting sphingosine to DMS. However, after substrate-specific experiments, we concluded that the enzymatic activity that we had isolated was sphingosine O-methyltransferase on C1 OH (Figure 5). We found enzymatic activity in the liver and kidney homogenates, but not in the brain, contrary to the previous report in which sphingosine N-methyltransferase was detected in the mouse brain under the same experimental conditions[6].
Methylation by SAM-dependent O-methyltransferases (OMT) is a common alkylation reaction that is observed in the metabolism of mammals and in natural product biosynthesis. Catechol O-methyltransferase (COMT) is a well-known enzyme that catalyzes the transfer of the methyl group of SAM to the phenolic group of catechol compounds, including norepinephrine and dopamine[20]. In mammals, COMT is distributed throughout various organs. In plant biosynthesis[21], plant class I OMT are a group of low molecular weight (23–27 kDa), Mg2+-dependent enzymes that were initially characterized as caffeoyl CoA OMT and shown to play an important role in the methylation of guaiacyl residue precursors of lignin, like caffeoyl or 5-hydroxyferuloyl CoA[22]. The substitution of Mg2+ with Ca2+ or Zn2+ in the caffeoyl CoA OMT assays marginally affected the substrate turnover rate and the substrate preference[23]. O-methylation by an inducible chlorophenol O-methyltransferase was almost completely inhibited by several metal ions, including Cu2+, Hg2+, Zn2+, and Ag+, whereas the inhibitory effects of Ca2+, Cs+, Fe2+, and NH4+ were weak. In our experiments, sphingosine 1-O-methyltransferase activity was inhibited by ZnCl2, but not by MgCl2, CaCl2, CoCl2, or NiCl2, suggesting variable susceptibility to metal ions among OMT.
Igarashi and Hakomori found sphingosine N-methyltransferase activity in the mouse brain homogenate[6]. They used mouse brain and liver tissue homogenates to establish the N-methyltransferase enzymatic activity. However, we could not find N-methyltransferase activity in the rat brain homogenate. In contrast, we found sphingosine O-methylation on the C1 OH group of sphingosines in the rat kidney and liver homogenates. Currently, we can not explain why our results differ from the previous report, because we were not able to detect sphingosine N-methyltransferase activity in the mouse brain and liver homogenates also (data not shown).
We found the activity in the cytosol and membrane fractions of the kidney homogenate. This might imply that 2 isoforms of sphingosine 1-O-methyltransferase are present in the kidney. One is an integral protein in membrane fractions and the other is a cytosolic protein in the cytosol. This could be 2 separate gene products, or the same gene could be transcriptionally modified, such as splicing variants or at the post-translation stage, such as palmitoylation for the enzyme in the membrane fractions. Further investigation to identify the gene would elucidate truth. In the liver, we only detected the activity in membrane fractions, but not in the cytosol. This suggests that only 1 gene is expressed in the liver or the modification in the kidney does not happen in the liver.
In summary, we found sphingosine 1-O-methyltransferase activity in the rat liver and kidney homogenates is also responsible for the conversion of sphingosine to methylated sphingolipids. Further investigation is necessary to determine whether 1-O-methylsphingosine is present in the liver and kidney to elucidate the pathophysiological significance of 1-O-methyl-sphingosine and to identify the enzyme at the molecular level for further studies of its regulation and relationship with certain diseases.
References
- Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol 2003;4:397-407.
- Cuvillier O. Sphingosine in apoptosis signaling. Biochim Biophys Acta 2002;1585:153-62.
- Pettus BJ, Chalfant CE, Hannun YA. Ceramide in apoptosis: an overview and current perspectives. Biochim Biophys Acta 2002;1585:114-25.
- Maceyka M, Payne SG, Milstien S, Spiegel S. Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim Biophys Acta 2002;1585:193-201.
- Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep 2004;5:777-82.
- Igarashi Y, Hakomori S. Enzymatic synthesis of N,N-dimethyl-sphingosine: demonstration of the sphingosine: N-methyltransferase in mouse brain. Biochem Biophys Res Commun 1989;164:1411-6.
- Mano N, Oda Y, Yamada K, Asakawa N, Katayama K. Simultaneous quantitative determination method for sphingolipid metabolites by liquid chromatography/ionspray ionization tandem mass spectrometry. Anal Biochem 1997;244:291-300.
- Kobayashi T, Mitsuo K, Goto I. Free sphingoid bases in normal murine tissues. Eur J Biochem 1988;172:747-52.
- Hannun YA, Bell RM. Lysosphingolipids inhibit protein kinase C: implications for the sphingolipidoses. Science 1987;235:670-4.
- Igarashi Y, Hakomori S, Toyokuni T, Dean B, Fujita S, Sugimoto M, et al. Effect of chemically well-defined sphingosine and its N-methyl derivatives on protein kinase C and src kinase activities. Biochemistry 1989;28:6796-800.
- Merrill AH Jr, Nimkar S, Menaldino D, Hannun YA, Loomis C, Bell RM, et al. Structural requirements for long-chain (sphingoid) base inhibition of protein kinase C in vitro and for the cellular effects of these compounds. Biochemistry 1989;28:3138-45.
- Edsall LC, Van Brocklyn JR, Cuvillier O, Kleuser B, Spiegel S. -Dimethylsphingosine is a potent competitive inhibitor of sphingosine kinase but not of protein kinase C: modulation of cellular levels of sphingosine 1-phosphate and ceramide. Biochemistry 1998; 37: 12 892–8.
- De Jonghe S, Van Overmeire I, Poulton S, Hendrix C, Busson R, Van Calenbergh S, et al. Structure–activity relationship of short-chain sphingoid bases as inhibitors of sphingosine kinase. Bioorg Med Chem Lett 1999;9:3175-80.
- Endo K, Igarashi Y, Nisar M, Zhou QH, Hakomori S. Cell membrane signaling as target in cancer therapy: inhibitory effect of N,N-dimethyl and N,N,N-trimethyl sphingosine derivatives on in vitro and in vivo growth of human tumor cells in nude mice. Cancer Res 1991;51:1613-8.
- Okoshi H, Hakomori S, Nisar M, Zhou QH, Kimura S, Tashiro K, et al. Cell membrane signaling as target in cancer therapy II: Inhibitory effect of N,N,N-trimethylsphingosine on metastatic potential of murine B16 melanoma cell line through blocking of tumor cell-dependent platelet aggregation. Cancer Res 1991;51:6019-24.
- Kimura S, Kawa S, Ruan F, Nisar M, Sadahira Y, Hakomori S, et al. Effect of sphingosine and its N-methyl derivatives on oxidative burst, phagokinetic activity, and trans-endothelial migration of human neutrophils. Biochem Pharmacol 1992;44:1585-95.
- Zatz M, Dudley PA, Kloog Y, Markey SP. Nonpolar lipid methylation. Biosynthesis of fatty acid methyl esters by rat lung membranes using -adenosylmethionine. J Biol Chem 1981; 256: 10 028–32.
- Mato JM, Alemany S. What is the function of phospholipid N-methylation? Biochem J 1983;213:1-10.
- Lim HS, Park JJ, Ko K, Lee MH, Chung SK. Syntheses of sphingosine-1-phosphate analogues and their interaction with EDG/S1P receptors. Bioorg Med Chem Lett 2004;14:2499-503.
- Mannisto PT, Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev 1999;51:593-628.
- Joshi CP, Chiang VL. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol Biol 1998;37:663-74.
- Ye ZH, Kneusel RE, Matern U, Varner JE. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 1994;6:1427-39.
- Lukacin R, Matern U, Specker S, Vogt T. Cations modulate the substrate specificity of bifunctional class I O-methyltransferase from Ammi majus. FEBS Lett 2004;577:367-70.
