Effects of RNAi-mediated inhibition of aggrecanase-1 and aggrecanase-2 on rat costochondral chondrocytes in vitro1
Introduction
Healthy cartilage consists of chondrocytes embedded in an extracellular matrix (ECM). The chondrocytes synthesize the matrix molecules, which maintain the structural integrity and mechanical properties of the tissue. The key components of the ECM are type II collagen and aggrecan, which comprise up to 90% of the dry weight of healthy cartilage. The major ECM proteoglycan is aggrecan. Its core protein is substantially modified with chondroitin sulfate and keratan sulfate glycosaminoglycans, which serve to hydrate the cartilage tissue, providing compression and elasticity[1,2]. Thus the maintenance of aggrecan content in articular cartilage is critical to the function of the tissue[3,4].
Cartilage damaged by trauma or degenerative disease has limited intrinsic potential for repair, due to the lack of blood supply. In clinical practice, costocartilage is often transplanted to serve as scaffolding for cartilage reconstruction. As the source of autologous cartilage is limited and has high potential donor site morbidity, it is common practice to transplant allogenic cartilage instead[5–7].
Failure of transplanted cartilage or allogenic chondro-cytes is characterized by degradation of cartilage and immunological rejection. A major feature is the loss of aggrecan from the cartilage matrix[7,8]. Many studies have shown that degenerative joint diseases are characterized by cartilage ECM degradation, where loss of aggrecan, an aggregating proteoglycan, is an early event in the destruction of the articular cartilage[9,10]. This loss of aggrecan results in a decrease in the functional and structural integrity of the cartilage matrix. As a consequence, the tissue loses its capacity to resist compression under load, which eventually leads to irreversible mechanical destruction of the cartilage[10]. Thus aggrecan turnover is an important process for the maintenance of homeostasis of the cartilage matrix.
Aggrecan comprises several segments: the N-termi-nal globular domains (G1 and G2), a long central glycosaminoglycan attachment region, and the C-terminal globular domain (G3)[11]. Aggrecan monomers interact with hyaluronic acid through the G1 domain to form aggregates of high molecular weight, which serve to retain aggrecan within the cartilage matrix[12,13].
A disintegrin and metalloproteinase with thrombospondin motifs(ADAMTS) are proteolytic enzymes that cleave aggrecan, disrupting cartilage homeostasis and weakening its biomechanical properties. Fragments generated by cleavage at specific sites in the core protein of aggrecan have been well-described and are detectable in the cartilage from cultured human cartilage explants[14]. Several members of the ADAMTS family possess some degree of aggrecanolytic activity in vitro, including ADAMTS-1, -4, -5, -8, -9, and -15[15–19]. However, ADAMTS-4 (aggrecanase-1) and ADAMTS-5 (aggrecanase-2) have the highest specific activity for aggrecan cleavage in vitro and are generally recognized as most likely to predominate in pathological processes involving the degradation of aggrecan in cartilage[20]. Thus the inhibition or inactivation of aggrecanase-1 and aggrecanase-2 would be expected to reduce or arrest the destruction of aggrecan.
In vitro, in cartilage cells and tissue culture systems, aggrecanase activity is upregulated by pro-inflammatory cytokines such as interleukin (IL)-1 and TNF-β and is therefore implicated in the initiation/progression of cartilage degeneration[21]. Thus strategies that inhibit aggrecanase activity should improve the viability of cartilage allografts by optimizing the longevity of the cartilage matrix and protecting the chondrocytes from the host’s immune system.
RNAi is a useful tool for identifying the role of different genes in disease progression. The method involves post-transcriptional gene silencing, in which dsRNA induces the degradation of homologous transcripts[22–24]. The introduction of 19–23 base pair RNA duplexes into mammalian cells leads to the specific degradation of target genes without arousing interferon responses[25,26]. Thus RNAi can be used in cultured cells, including mammalian cells, primary cells, embryonic stem cells, and in animals[27]. In vector-based siRNA, plasmid vectors are transiently transfected into the cell to downregulate gene expression.
In this study, we attempted to construct plasmid vectors of shRNA targeting aggrecanase-1 and aggrecanase-2, transiently transfected into cultured rattus costochondral chondrocytes, in order to study the effects of aggrecanase inhibition on gene expression and ECM stability in vitro.
Materials and methods
Materials and reagents Sprague–Dawley (SD) rats were obtained from the animal center of Xi’an Jiao Tong University (Xi’an, Shaanxi, China). The PshRNA–GFP lentivector kit was purchased from Kangcheng (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM), Lipofectamine 2000, and TRIzol were from Invitrogen (Carlsbad, CA, USA). Fetal bovine serum (FBS) was from BioInd (Kibbutz Beit Haemek, Israel). IL-1β was from Reprotech (Rehovot, Israel). The reverse first-strand cDNA synthesis kit was from MBI Fermentas (Glen Burnie MD, USA). The mouse polyclonal antibody to β-actin (rat) and the goat antibody to aggrecan (rat) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The aggrecan-specific NITEGE373 neoepitope rabbit polyclonal antibody (rat) was from IBEX (Montréal, Québec, Canada). The mouse polyclonal antibody against collagen II (rat) was from Lab Vision (Fremont, CA, USA). Goat antimouse immunoglobulin G (IgG)/horseradish peroxidase (HRP), goat antirabbit IgG/HRP, and rabbit antigoat IgG/HRP were from Zhongshan (Beijing, China). The Supersignal West Pico chemiluminescent kit was from Pierce (Rockford, IL, USA).
ShRNA template design and preparation Three siRNA sequences specific to each type of gene were chosen from the rat aggrecanase-1 (GenBank Accession N

Full table
Isolation and culture of rat costochondral chondrocytes All procedures were approved by the Animal Care and Use Committee of Xi-an Jiao Tong University. The ribs medial to the costochondral junction were dissected from anesthetized 1-month-old SD rats with the sternum still attached[28]. The dissected rib cages were stored in Hanks’ buffered salts solution until the required number of rats had been dissected. The rib cartilages were then sliced into 1 mm3 cubes and digested with 0.05% hyaluronidase (Sigma, St Louis, MO, USA) for 0.5 h at 37 °C, 0.25% trypsin (Gibco, Carlsbad, CA USA) for 0.5 h at 37 °C, and subsequently, 0.2% collagenase II (Sigma, USA) overnight at 37 °C with moderate agitation. After digestion, the released cells were passed through a 100 μm nylon mesh to remove undigested fragments. The filtrate was centrifuged at 1 300×g for 5 min and the cell pellets were resuspended in culture medium at the required density for direct seeding. The culture medium consisted of DMEM supplemented with 584 mg/L glutamine, 50 mg/L L-ascorbic acid, 5×104 U/L penicillin, 50 mg/L streptomycin sulfate, 10 mmol/L HEPES, and 10% fetal bovine serum. The chondrocytes were cultured in 25 cm2 cell culture flasks at a final density of 1×105 cells/cm2 as monolayers at 37 °C in a humidified 5% (v/v) CO2 incubator. The medium was changed every 2 d. The chondrocytes were passaged on approximately the seventh day when the cells were 90% confluent.
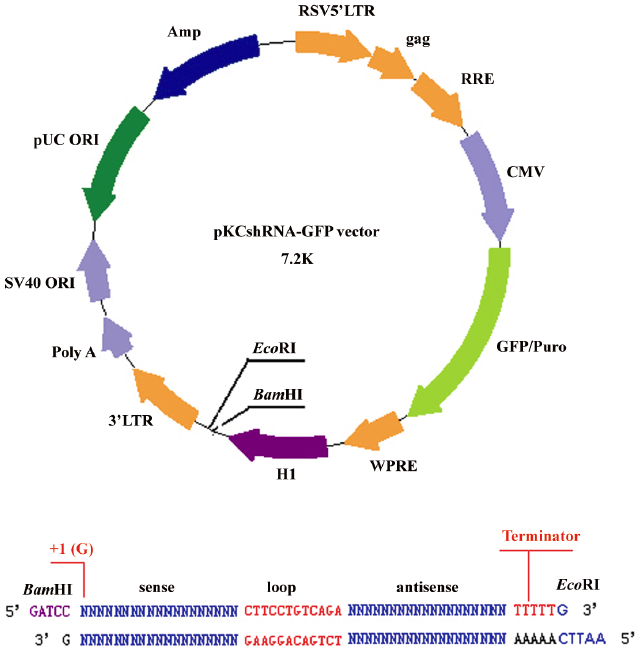
Construction of plasmid vectors and transfection The synthesized shRNA sequences were cut by BamHI and EcoRI. The expression vector (Figure 1) was digested with BamHI and EcoRI and gel purified. The synthesized shRNA sequences were linked into the BamHI–EcoRI restriction sites of the vector by the T4 DNA ligase. Clones were verified by sequencing[29].
The cultured passage 1 chondrocytes were seeded at 1×106 per well in 6-well plates. After 1 d the cells were transfected with the expression plasmids, using Lipofectamine 2000 reagent according to the manufacturer’s instructions at 80%–90% confluency. After 6 h of transfection, the complexes were removed from the 6-well plates. The cells were washed with phosphate-buffered saline (PBS), and 2 mL fresh medium containing 10% FBS was added to the plates. After 2 or 3 d, chondrocytes were harvested from 3 independent wells and used for the following analyses, each of which was performed at least 3 times. The controls and negative controls were analyzed in parallel.
Observation of cell morphology and growth velocities After 6 h of transfection, the cells were transferred to 12-well plates (5×104 cells per well) in triplicate. The chondrocyte morphology was observed daily for 7 d using an inverted microscope, and the cells were counted by a hemocytometer (Cole-Parmer, Vernon Hills, IL, USA) in triplicate to generate growth curves.
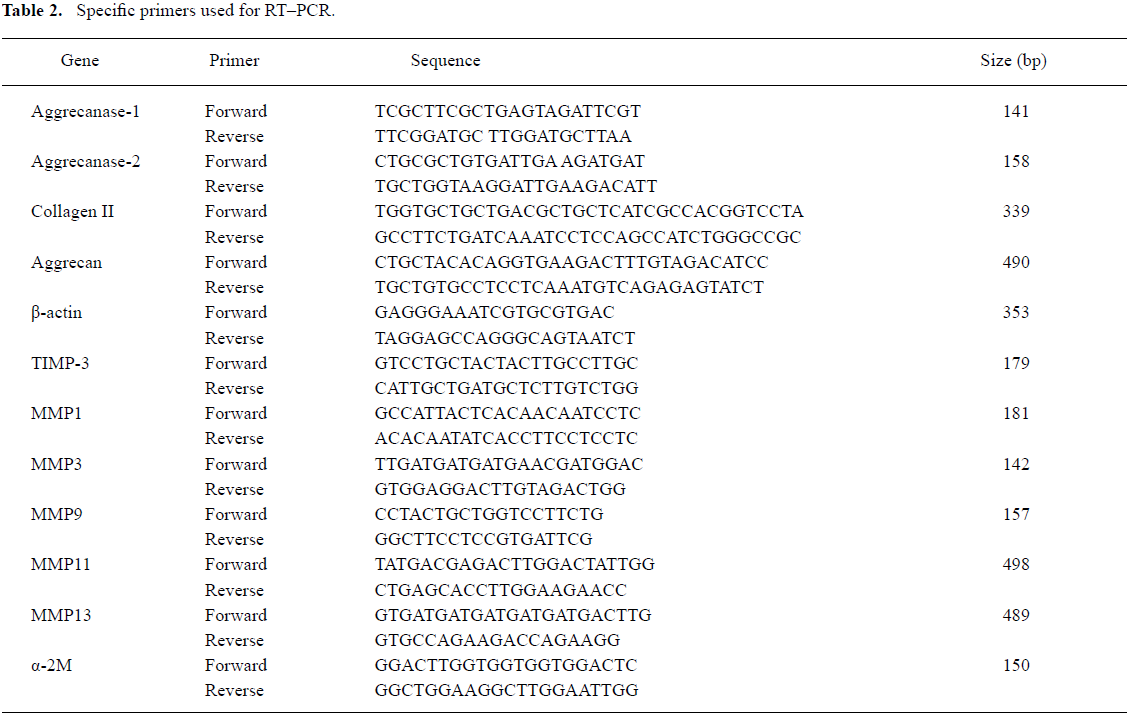
RT–PCR The total RNA was extracted from the chondrocytes after 48 h of transfection using TRIzol reagent according to the manufacturer’s protocol. First-strand cDNA was synthesized using the reverse RT–PCR reagent kit, according to the manufacturer’s instructions. The specific primers used for RT–PCR are shown in Table 2. β-actin was amplified as an internal control for normalization. Aliquots of each RT sample were assessed for β-actin expression following 35 PCR cycles. PCR products were separated on 1.5% agarose gels and visualized by ethidium bromide staining. The images were analyzed by the GEL DOC 2000 system (Bio-Rad, Hercules, CA, USA). The relative gene expression level (%)=gene band density/β-actin band density.
Full table
Cytokine induction After the 6 h of shRNA treatment, the culture medium was replaced with DMEM containing 0.5% FBS. One day later, chondrocytes in 3 wells were incubated in 0.5% FBS with rat IL-1β (100 ng/mL) for another 24 h, while chondrocytes in 3 additional wells were exposed to the same conditions without IL-1β. The cells were then processed for RNA extraction. Control chondrocytes were treated and analyzed in parallel.
Immunohistochemical staining The cells were fixed in 4% paraformaldehyde for 30 min and treated with methanol and 0.1% Triton X-100 to achieve cell membrane and nuclear membrane permeability. Non-specific binding was blocked by incubation with normal goat serum for 30 min. The chondrocytes were incubated with 1:200 mouse anti-rat collagen II or a goat antirat aggrecan antibody overnight at 4 °C and then washed. A universal biotinylated secondary antibody was applied for 30 min at room temperature. After incubation, the plates were washed thoroughly with PBS containing 1% bovine serum albumin (BSA) and incubated with horseradish peroxidase-labeled streptavidin (ABC kit; Vector Laboratories, Elk Grove CA, USA). This process was followed by reaction with diaminobenzidine (Sigma, USA). The negative control was treated without a primary antibody. Representative cells were photographed under an Olympus IX 70 microscope (Tokyo, Japan). The gray density was analyzed by an image analysis system (Leica, Solmser Gewerbepark, Solms, Germany).
Western blotting To detect aggrecanase-generated aggrecan fragments with NITEGE373 neoepitope antibodies, after the 72 h shRNA treatment, cells from a single plate were homogenized in lysis buffer (50 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 1% NP-40, and 0.1% SDS, supplemented with protease and phosphatase inhibitors). The proteins were separated on 4%–12% polyacrylamide gels containing 0.1% SDS and then transferred to nitrocellulose. After blocking for 4 h in blocking buffer (5% non-fat dried milk and 0.5% Tween-20 in Tris buffered solution (TBS) and separate incubation with the NITEGE373 neoepitope monoclonal antibody (1:200) and an antirat β-actin mouse polyclonal antibody (1:1000) overnight at 4 °C, the blots were washed 3 times with TBST (0.5% Tween in TBS) and incubated for 1 h with goat antirabbit IgG/HRP (1:7000) or goat antimouse IgG/HRP (1:8000) at room temperature, followed by washing 3 times with TBST. The signals were visualized with the enhanced chemiluminescence method and developed on X-ray film. The band density was measured by the GEL DOC 2000 system equipped with Quantity One software (Bio-Rad, USA) and normalized against the density of the respective housekeeping gene β-actin.
Statistical analysis The data were statistically evaluated by ANOVA and presented as mean±SD from 3 independent experiments. Probability values less than 0.05 were considered significant. All analyses were carried out using SPSS 10.0 statistical software (SPSS, Chicago, IL, USA).
Results
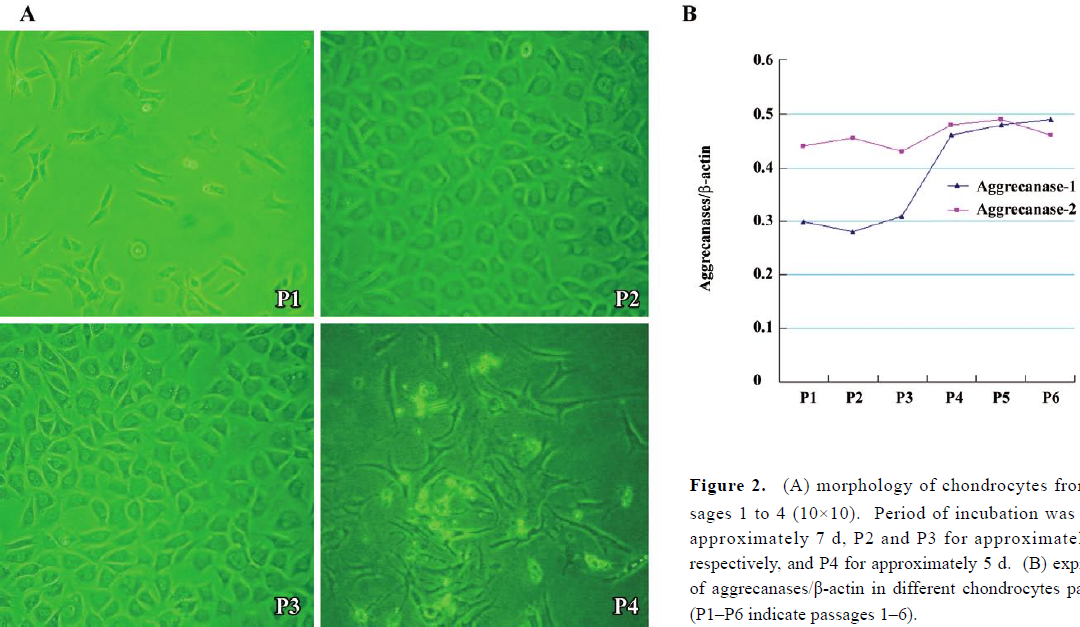
Expressions of aggrecanase-1 and aggrecanase-2 in costochondral chondrocytes The morphology of costochondral chondrocytes from passages 1–3 showed round, polygonal cells in a cobblestone pattern when cell density was high. In passage 4, the cells became long and fusiform in shape, resembling fibroblasts (Figure 2A).The mRNA of aggrecanase/β-actin (Figure 2B) showed that the aggrecanase-1 expression was low from passages 1–3 and then increased significantly in passage 4 compared to the other passages (P<0.05). The aggrecanase-1 expression then remained high, while the aggrecanase-2 expression showed no significant variations through the different passages.
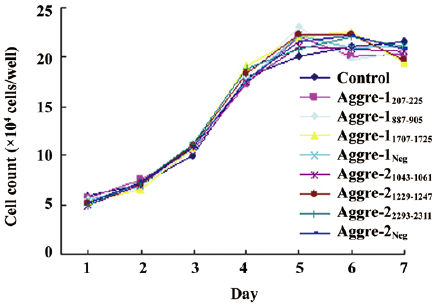
Observation of cell morphology and growth velocities RCC were transfected with plasmids expressing shRNA specific to aggrecanase-1 and aggrecanase-2, and the transfection rate of cells was approximately 60%. Cells treated with RNAi exhibited no significant changes in morphology or growth compared with the controls. In all cases, RCC were polygonal or round in shape and reached confluence after 2–3 d. Cells grew and formed a cobblestone pattern. A growth curve analysis suggested that cells reached the log phase after 2 d, and peak growth by 5 days (Figure 3). No statistically significant differences were observed between each transfected group and the untransfected control, or among the transfected groups (P>0.05).
Knockdown of aggrecanase-1 and aggrecanase-2 expression in chondrocytes by RNAi To validate the activity of each shRNA designed to inhibit aggrecanase-1 and aggrecanase-2 expressions, several plasmids were transfected into chondrocytes, and the degrees of gene knockdown were assessed by RT–PCR. Only aggre-1887–905 or aggre-11707–1725 induced a significant reduction in the gene expression of aggrecanase-1 48 h post-transfection. These 2 plasmids were capable of specifically inhibiting the gene expression (46.15% of control levels for aggre-1887–905, P<0.01, and 49.14%, P<0.01 for aggre-11707–1725, respectively; Figure 4A, 4B). When treated with RNAi plasmids, the aggrecanase-2 expression was significantly inhibited by aggre-21043–1061 or aggre-21229–1247. Compared to the control levels, the gene expression was 60% for aggre-21043–1061 (P<0.05), and 71.64% for aggre-21229–1247 (P<0.05) (Figure 4C, 4D). ShRNA-aggre-1887–905 and ShRNA-aggre-21043–1061 emerged as the most potent sequences for aggrecanase-1 and aggrecanase-2. Other shRNA resulted in a moderate to very poor reduction of aggrecanase-1 and aggrecanase-2 gene expressions (Figure 4). In contrast, the expressions of aggrecanase-1 and aggrecanase-2 in the negative control changed only negligibly (97.20% and 96.69%, respectively), indicating that the mediated knockdown was highly specific.
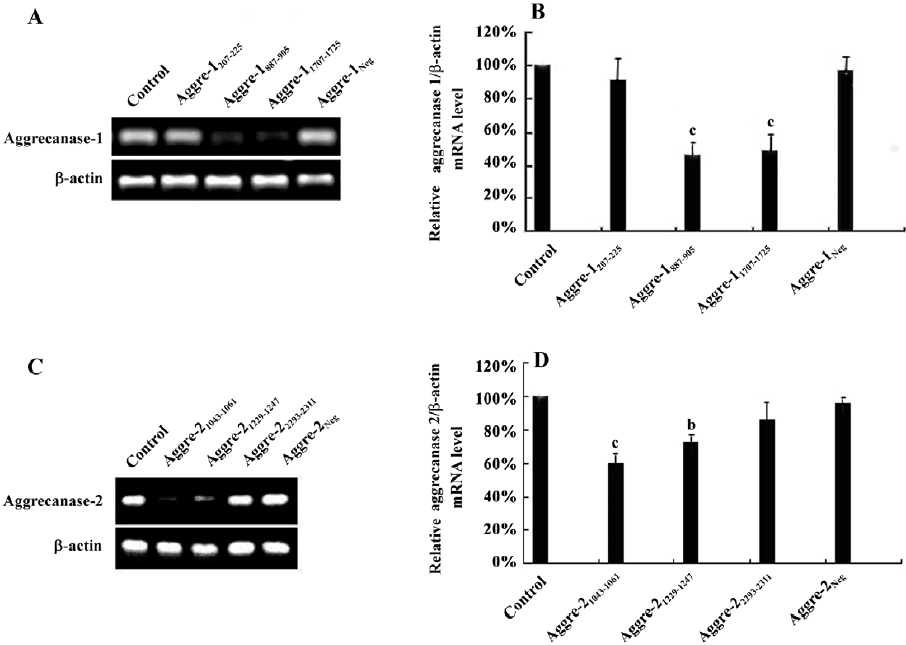
Effect of RNAi on other chondrocytic ECM-related genes To observe the effects of the inhibition of aggrecanase-1 and aggrecanase-2 on chondrocytic ECM-related genes, we isolated the RNA after 48 h of shRNA plasmid transfection. RT–PCR showed that the α-2-macroglobulin (α-2M) expression level was increased by the shRNA specific for aggrecanase-1. There were significant differences between aggre-1887–905 and aggre-11707–1725 groups compared to the untreated control (P<0.01; Figure 5A, 5B). There were no significant differences in the α-2M expression among the RNAi of the aggrecanase-2 groups (P>0.05; Figure 5C, 5D). The tissue inhibitor of metalloproteinase-3 (TIMP-3) mRNA levels did not differ among the groups of shRNA for aggrecanase-1 or aggrecanase-2. Moreover, no differences were observed in the mRNA expressions of matrix metalloproteinase
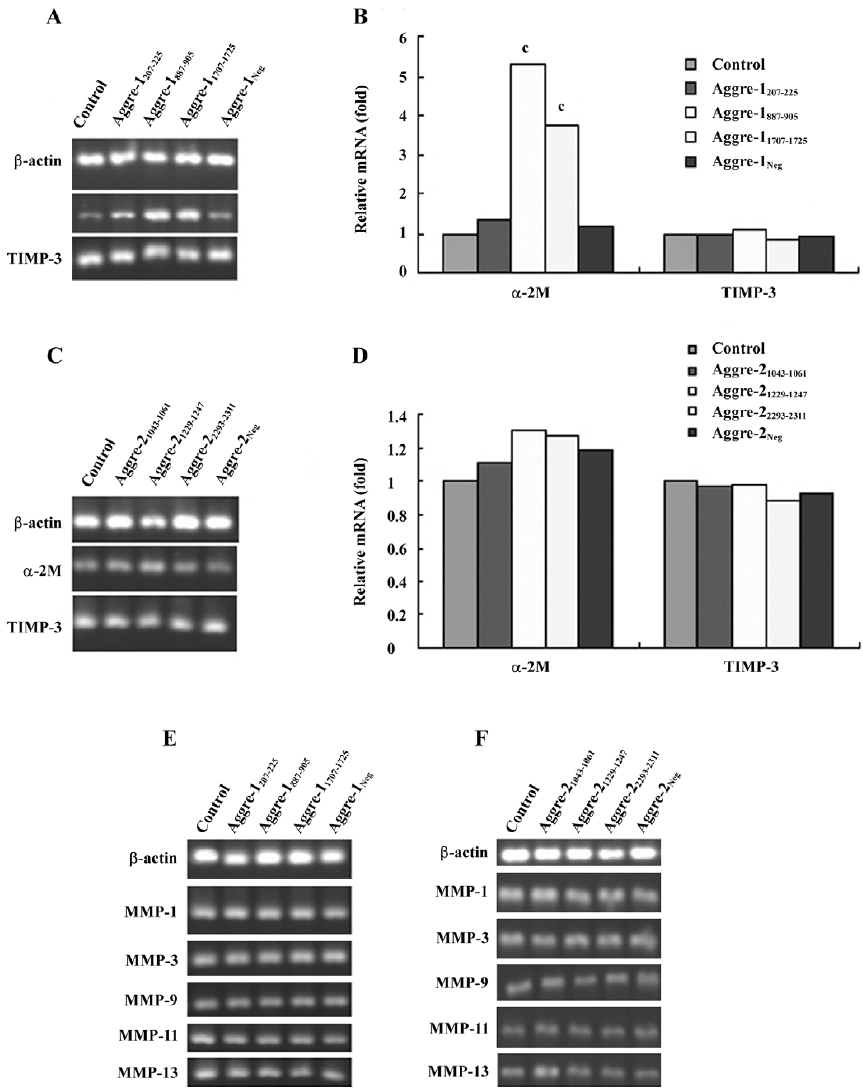
Effects of cytokine induction on chondrocytes The RT–PCR analysis (Figure 6) shows that the aggrecan mRNA levels of the IL-1β-treated group were significantly lower than those of the control group (P<0.01). Using the most potent RNAi sequence identified for aggrecanase-1 and aggrecanase-2, we found that the chondrocyte levels of mRNA for aggrecan increased by 35%–40% relative to the corresponding levels observed in cells treated only with IL-1β (P<0.05).
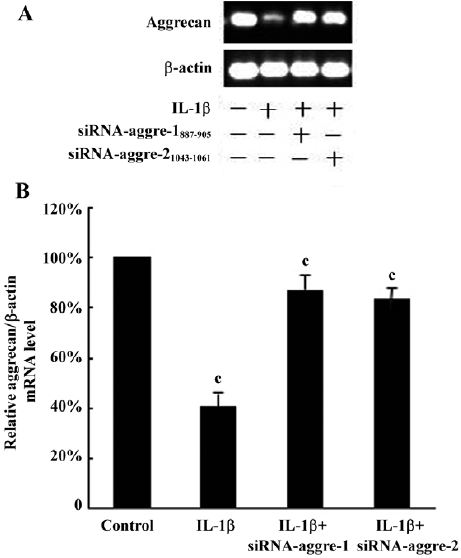
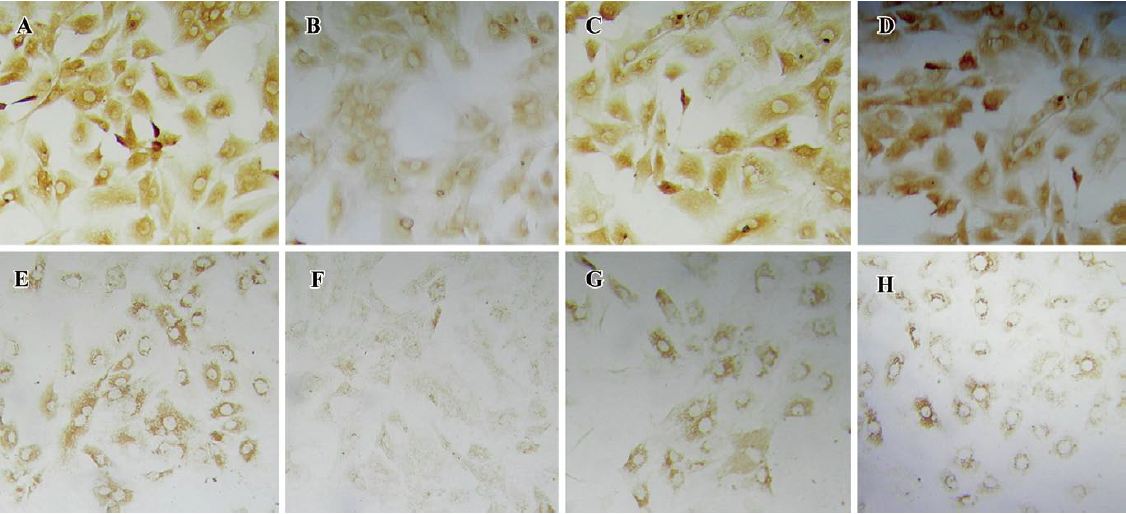
The immunohistochemical analysis found the expression of aggrecan throughout the cellular component of each group. After incubation with IL-1β (100 ng/mL) for 24 h, the positive staining for aggrecan decreased significantly compared with the control group (P<0.05; Figure 7B). Incubation with IL-1β+shRNA-aggre-1887–905 or shRNA-aggre-21043–1061 yielded no significant differences in the aggrecan content (Figure 7A, 7C, 7D) compared with the control group (P>0.05). At the same time, incubation with IL-1β significantly decreased the collagen II content (Figure 7F) compared with the control group (P<0.05). After incubation with IL-1β+shRNA-aggre-1887–905 or shRNA-aggre-21043–1061, the collagen II content (Figure 7G, 7H) of the chondrocytes increased significantly compared with the IL-1β group (P<0.05).
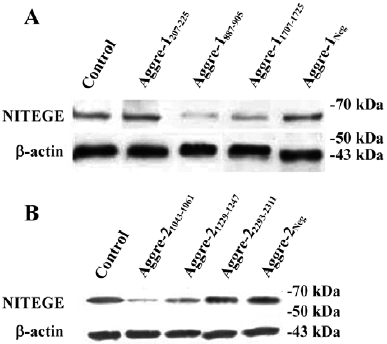
Effects of aggrecanase-1 and aggrecanase-2 gene silencing on aggrecan degradation in chondrocytes We next correlated the effects on glycosaminoglycan (GAG) release with the effects on the release of the NITEGE373 aggrecan neoepitopes, which are generated when aggrecanases cleave the aggrecan core protein at the site. There was a significant decrease in the release of the NITEGE373 aggrecan neoepitope, as detected by the Western blot analysis (Figure 8). Compared with the control levels, the NITEGE373 levels of aggre-1207–275 or aggre-1887–905 relatively decreased significantly, as did the NITEGE373 levels of aggre-21043–1061 and aggre-21229–1247. Other shRNA against aggrecanase-1 and aggrecanase-2 had no significant effects on the NITEGE373 levels. These results were consistent with the RT–PCR analysis showing that compared with untransfected controls, several plasmids expressing shRNA had depressed the aggrecanase-1 and aggrecanase-2 mRNA levels.
Discussion
Allogenic cartilage transplants may fail due to immunological rejection, resulting in the absorption of cartilage, which compromises the long-term viability of the tissue[8]. Cartilage is an avascular tissue and its ECM does not allow permeability of molecules greater than 60 kDa[8]. Therefore, the long-term survival of allogenic transplants of whole cartilage fragments may be explained by sequestration of chondrocyte-associated histocompatibility antigens by the matrix, preventing immunization of the host. Because of the matrix, cartilage is a poor antigen tissue. While maintenance of the stability of the matrix is important, there is also a need to address the issue of immunological rejection of cartilage transplantation.
The loss of aggrecan from the ECM is not only a key feature in failure of allogenic cartilage transplants[4–7], but also an early event in the destruction of articular cartilage in degenerative joint disease[9,10]. Aggrecanase-1 and/or aggrecanase-2 are the enzymes responsible for aggrecan cleavage during cytokine-induced cartilage degradation[30–32]. Thus inhibition or inactivation of aggrecanase-1 and aggrecanase-2 activity would be expected to reduce or arrest the destruction of aggrecan.
In our previous studies, we reported the marked depletion of aggrecan in cartilage ECM, particularly in the first month after cartilage transplantation surgery[4,6,7]. Early aggrecan loss was also observed after allogenic chondrocyte transplantation[8]. The pattern of matrix degradation resembled that of degenerative joint diseases.
The present study sought to address several problems associated with allogenic cartilage and chondrocyte transplants. We applied shRNA-based gene silencing to rat costochondral chondrocytes and used the technique to further elucidate the roles of specific aggrecanases in cartilage aggrecan degradation. The results suggest that the inhibition of aggrecanase-1 and aggrecanase-2 by RNAi can reduce aggrecan degradation.
Once the chondrocytes had been successfully cultured in vitro, study of cartilage matrix progressed rapidly. However, some issues remain unresolved, such as the dedifferentiation, which occurs in cultured chondrocytes in vitro. As the number of passages increases, the cells undergo a change in phenotype and the morphology becomes more fibroblast-like. Dedifferentiated chondrocytes lose their ability to form a matrix; they predominantly synthesize type-1 collagen. There is associated downregulation of both aggrecan and type II collagen, the major protein produced by chondrocytes in cartilage[28,33]. Thus maintenance of the chondrocyte phenotype during a prolonged monolayer culture, delaying dedifferentiation, is crucial to chondrocyte transplantation and tissue engineering.
In the present study, the transition to fibroblast-like morphology was observed at passage 4. At this passage, the mRNA level of aggrecanase-1 increased more significantly than in previous passages, while aggrecanase-2 levels remained unchanged. The results suggest a possible relationship between the aggrecanase-1 mRNA level and chondrocyte dedifferentiation[34]. Chondrocytes from passages 1–2 were harvested for use in this study.
Gene silencing using double-stranded siRNA is widely used to examine candidate gene functions in a range of normal physiological and pathological processes. This technology, which causes the specific degradation of mRNA with sequences homologous to the introduced duplexes, has been applied to many cell types and tissues. Electroporation of siRNA into the knee joints of live mice and rats has been shown to ameliorate inflammation in collagen-induced arthritis through the targeted suppression of TNF-α[35,36]. SiRNA transfected into human chondrocytes and explants has been shown to mitigate the loss of aggrecan by the inhibition of ADAMTS[37].
In the present study, using RCC as the object, both aggrecanase-1 and aggrecanase-2 plasmids expressing shRNA treatments clearly inhibited aggrecanase-1 and aggrecanase-2 mRNA levels. The most potent RNAi sequences identified for aggrecanase-1 and aggrecanase-2 were shRNA-aggre-1887–905 and shRNA-aggre-21043–1061. However, in the study the inhibition rate of aggrecanases was relatively lower compared with other RNAi studies, which may be due to the low transfection rate of cells. Meanwhile, the inhibition of aggrecanase-1 and aggrecanase-2 did not adversely affect chondrocyte morphology or growth velocities. The results suggest that RNAi technology does not harm chondrocytes.
The effects on aggrecan loss were correlated with the effects on the release of the NITEGE373 aggrecan neoepitopes, which are specific for the site at which aggrecanase cleaves the aggrecan core protein[38]. In this study, the effects of RNAi aggrecanase-1 and aggrecanase-2 on aggrecan loss were analyzed by Western blotting. The NITEGE373 protein level decreased significantly following the inhibition of aggrecanase-1 and aggrecanase-2. Thus the decrease in the release of aggrecan NITEGE373 neoepitope associated with gene silencing of aggrecanase-1 and aggrecanase-2 corresponded with the decrease in aggrecan degradation.
As a naturally occurring inhibitor of aggrecanase-1, α-2M is found at high levels in synovial fluid and inhibits aggrecanase-1 with a second-order rate constant in the order of 106 (mol/L)-1·s-1 [39]. In the present study, using RT–PCR to observe the effect of RNAi on other ECM genes, α-2M mRNA levels increased significantly after inhibition of aggrecanase-1, but were unaffected by the inhibition of aggrecanase-2. It has recently been demonstrated that aggrecanase-1 is not exclusively a glutamyl endopeptidase and not limited to cleavage of proteoglycans[40]. Aggrecanase-1 cleaves α-2M within the “bait region” following Met690. In addition, this cleavage of the macroglobulin molecule is not dependent on the thrombospondin-1(TSP-1) motif, as a truncated form of the enzyme213–431 lacking the C-terminal domains431–837 is equally active against α-2M[39]. The activity of both ADAMTS-4 and ADAMTS-5 was inhibited by α-2M in a concentration-dependent manner[40]. As stated earlier, aggrecanase cleaves α-2M within the ‘bait region” at Met690/Gly691, representing a novel proteinase cleavage site within α-2M and a novel site of cleavage for ADAMTS-4 and ADAMTS-5. The cleavage of α-2M within the bait region at Met690 appears to be unique for the aggrecanases[40]. Thus the inhibition of aggrecanase-1 may result in an increase in α-2M mRNA levels. TIMP-3 is an aggrecanase inhibitor[41]. The present study found no difference of inhibition of aggrecanase-1 and aggrecanase-2 on TIMP-3 or on other MMP tested in the study.
In vivo, the tissue destruction associated with cartilage transplantation may be driven not only by a single cytokine or cytokine combination such as that used to stimulate breakdown in vitro, but is also likely to involve other aberrant signalling processes, such as those induced by chondrocyte binding of proteolytically derived matrix fragments[5,42]. In the present study, we observed the effects of cytokine induction on chondrocytes. RT−PCR and immunohistochemical staining found that treatment with IL-1β significantly reduced the aggrecan and collagen content. After inhibition of aggrecanase-1 and aggrecanase-2 by RNAi technology, aggrecan and collagen increased significantly. These results demonstrated gene phenotype recovery by the chondrocytes, which showed greater resistance to IL-1β after the inhibition of aggrecanase-1 and aggrecanase-2. It has previously been shown, in osteoarthritic chondrocytes, that silencing p16INK4a by shRNA was effective in maintaining the chondrocytic phenotype as culture proliferation increased, thus delaying dedifferentiation[43]. Aggrecans are polymers occurring in chondrocytic ECM. In vitro, in cartilage and tissue culture systems, they are entrapped in the collagen frame of the tissue. We therefore speculated that the aggrecan protected the cartilage from collagen loss and facilitated gene phenotype recovery of the chondrocytes.
The present study further found that the mRNA of aggrecanase-1 increased as the chondrocytes aged, thus the issue of chondrocyte dedifferentiation may be addressed through the study of aggrecanase-1. Whether dedifferentiation can be postponed by the inhibition of aggrecanase-1 and aggrecanase-2 warrants further investigation.
RNAi technology is a useful tool for studying cartilage. In future studies, plasmids expressing validated sequences of aggre-1887–905 or aggre-21043–1061 may be used to clone stable expression aggrecanase-1 and aggrecanase-2-inhibited chondrocytes. The results of the present study indicate that further investigation is warranted to determine whether gene silencing of aggrecanase-1 and aggrecanase-2 can reduce immunological rejection and cartilage degradation associated with cartilage transplantation.
Acknowledgement
The excellent technical assistance of Jian-feng SHI (Research Center of Stomatological Hospital) is gratefully acknowledged.
References
- Hardingham TE, Fosang AJ. Proteoglycans: many forms and many functions. FASEB J 1992;6:861-70.
- Vertel BM. The ins and outs of aggrecan. Trends Cell Biol 1995;5:458-64.
- Kouji K, Yasunori O, Hiroto K, Hiroyuki N, Masayuki M, Hiroshi O, et al. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett 2000;478:241-5.
- Zheng XM, Yang ZQ, Hou CQ, Wang Y, Chang XF, Xu YC, et al. The effects of cartilage matrix on cartilage implantation mechanisms. Chin J Plast Surg 1999;15:74-5. (in Chinese).
- Yang ZQ, Hou CQ, Chang XF, Wang Y, Xu YC, Zheng XM. Experimental observation on the dynamic changes of cartilage matrix in homografts. Chin J Aesthetic Med 1997;6:14-6. (in Chinese).
- Hou CQ, Yang ZQ, Chang XF, Wang Y, Xu YC, Yang XP, et al. An experimental study on changes in collagen content after homologous cartilage implantation. Chin J Plast Surg 1995;11:274-6. (in Chinese).
- Zheng XM, Yang ZQ, Hou CQ, Feng B, Wang Y, Xu YC. Experimental observations of the quantitative change of proteoglycan in homogenous cartilage graft in the guinea pig. Chin J Plast Surg 1995;11:129-31. (in Chinese).
- Romaniuk A, Malejczyk J, Kubicka U, Hyc A, Olszewski W, Moskalewski S. Rejection of cartilage formed by transplanted allogeneic chondrocytes: evaluation with monoclonal antibodies. Transplant Immunol 1995;3:251-7.
- Mankin HJ, Lippiello L. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. J Bone Joint Surg Am 1970;52:424-34.
- Lohmander LS, Neame PJ, Sandy JD. The structure of aggrecan fragments in human synovial fluid evidence that aggrecanase mediates cartilage degradation in inflammatory joint disease, joint injury, and osteoarthritis. Arthritis Rheum 1993;36:1214-22.
- Doege K, Sasaki M, Horigan E, Hassell JR, Yamada Y. Complete primary structure of the rat cartilage proteoglycan core protein deduced from cDNA clones. J Biol Chem 1987; 262: 17 757–67.
- Heinegård D, Hascall VC. Aggregation of cartilage proteoglycans.3. Characteristics of the proteins isolated from trypsin digests of aggregates. J Biol Chem 1974;249:4250-6.
- Hardingham TE. The role of link-protein in the structure of cartilage proteoglycan aggregates. Biochem J 1979;177:237-47.
- Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella M. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem 2002; 277: 22 201–8.
- Collins-Racie LA, Flannery CR, Zeng W, Corcoran C, Annis-Freeman B, Agostino MJ, et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol 2004;23:219-30.
- Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, . Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4). J Biol Chem 2000; 275: 18 566–73.
- Rodriguez-Manzaneque J, Westling J, Thai S, Luque A, Knauper V, Murphy G, et al. ADAMTS-1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem Biophys Res Commun 2002;293:501-8.
- Tortorella MD, Liu RQ, Burn T, Newton RC, Arner E. Characterization of human aggrecanase 2 (ADAMTS5): substrate specificity studies in comparison with aggrecanase 1 (ADAMTS4). Matrix Biol 2002;21:499-511.
- Somerville RP, Longpre J, Jungers K, Engle J, Ross M, Evanko S, et al. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans gon-1. J Biol Chem 2003;278:9503-13.
- Pratta MA, Tortorella MD, Arner EC. Age related changes in aggrecan glycosylation affect cleavage by aggrecanase. J Biol Chem 2000; 275: 39 096–102.
- Arner EC, Pratta MA, Trzaskos JM, Decicco CP, Tortorella MD. Generation and characterization of aggrecanase. A soluble, cartilage-derived aggrecan degrading activity. J Biol Chem 1999;274:6594-601.
- Hannon G J. RNA interference. Nature 2002;418:244-51.
- Caplen NJ. Gene therapy progress and prospects: down-regulating gene expression: the impact of RNA interference. Gene Ther 2004;11:1241-8.
- Mello CC, Conte D Jr. Revealing the world of RNA interference. Nature 2004;431:338-42.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:428-9.
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA 2001;98:9742-7.
- Gou D, Jin N, Liu L. Gene silencing in mammalian cells by PCR based short hairpin RNA. FEBS Lett 2003;548:113-8.
- Wang ZH, Li GG, Yang ZQ, Wang L, Li LX, He XJ. Culture of rat costochondral chondrocytes and effect of aging on matrix catabolism of cultured chondrocytes. J Tissue Eng Reconstr Surg 2007;5:312-6. (in Chinese).
- Wang ZH, Yang ZQ, He XJ, Li GG, Wang L, Li LX. Construction and sequencing of recombinant lentiviral plasmid affecting rattus aggrecanase-1, 2 gene by RNA interfering. Chin J Aesth Med 2007;16:1679-82. (in Chinese).
- Sandy JD, Flannery CR, Neame PJ, Lohmander LS. The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu 373-Ala 374 bond of the interglobular domain. J Clin Invest 1992;89:1512-6.
- Tortorella MD, Malfait AM, Deccico C, Arner E. The role of ADAMTS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthritis Cartilage 2001;9:539-52.
- Arner EC, Pratta MA, Decicco CP, Xue CB, Newton RC, Trzaskos JM, et al. Aggrecanase a target for the design of inhibitors of cartilage degradation. Ann NY Acad Sci 1999;30:92-107.
- Stokes DG, Liu G, Coimbra IB, Piera-Velazquez S, Crowl RM, Jimenez SA. Assessment of the gene expression profile of differentiated and dedifferentiated human fetal chondrocytes by microarray analysis. Arthritis Rheum 2002;46:404-19.
- Li Q, Lu JH, Liu W, Xia WR, Liu DL, Cui L, et al. The effects of aging on aggrecan catabolism in cultured chondrocytes. Chin J Clin Rehab 2003;7:216-8. (in Chinese).
- Schiffelers RM, Xu J, Storm G, Woodle MC, Scaria PV. Effects of treatment with small interfering RNA on joint inflammation in mice with collagen-induced arthritis. Arthritis Rheum 2005;52:1314-8.
- Inoue A, Takahashi KA, Mazda O, Terauchi R, Arai Y, Kishida T, et al. Electro-transfer of small interfering RNA ameliorated arthritis in rats. Biochem Biophys Res Commun 2005;336:903-8.
- Song RH, Tortorella MD, Malfait AM, Alston JT, Yang ZY, Elizabeth CA, et al. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum 2007;56:575-85.
- Arner EC. Aggrecanase-mediated cartilage degradation. Curr Opin Pharmacol 2002;2:322-9.
- Tortorella MD, Arner EC, Hills R, Gormley J, Fok K, Pegg L, . Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J Biol Chem 2004; 279: 17 554–61.
- Micky DT, Elizabeth CA, Robert H, Jennifer G, Kam F, Lyle P, et al. ADAMTS-4 (aggrecanase-1): N-Terminal activation mechanisms. Arch Biochem Biophys 2005;444:34-44.
- Gakuji H, Takanori A, Hiroyuki N, Kazuhiko T, Yasunori O. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4). FEBS Lett 2001;494:192-5.
- Homandberg G. Potential regulation of cartilage metabolism in osteoarthritis by fibronectin fragments. Front Biosci 1999;15:D713-30.
- Zhou HW, Lou SQ, Zhang K. Recovery of function in osteoarthritic chondrocytes induced by p16INK4a-specific siRNA in vitro. Rheumatology 2004;43:555-68.