Biotransformation of metoprolol by the fungus Cunninghamella blakes-leeana
Introduction
Metoprolol, 1-isopropylamino-3-(4-[2-methoxyethyl]-phenoxy)-2-propanol, is a β1 selective aryloxypropanolamine used in treatment of cardiovascular disorders such as hyper-tension, arrhythmia, and heart failure. The drug is a lipophilic adrenoceptor antagonist (β-blocker) with a short half life (3–4 h)[1]. It is therapeutically used as a racemic mixture.
Metoprolol is mainly eliminated by the hepatic oxidative metabolism and it undergoes extensive first pass metabolism with about 95% of the dose being metabolized in humans. Metoprolol is metabolized to a large degree by cytochrome P-450 2D6 which is polymorphic in the human population. The appearance of major metabolites O-desmethyl-meto-prolol, metoprolol acid, and hydroxymetoprolol varies depending on the oxidation phenotype[2–4].
The identification of metabolites from mammalian sources may be hindered by insufficient quantities of material. The concept of using microorganisms, particularly the filamentous belonging to the genus Cunninghamella, as models of mammalian metabolism has been well documented[5–7]. The advantages of a microbial system as an in vitro model for drug metabolism include its low cost, ease of handling, scale-up capacity, and potential to reduce the use of mammals. A microbial system also provides enough putative metabolites under milder conditions than those required by chemical systems. Thus, fermentation is used to scale up the synthesis of metabolites for structural confirmation by nuclear magnetic resonance (NMR)[8].
Although the metabolism of metoprolol in mammals has previously been studied, to our knowledge, no report has been published on the potential of filamentous fungi to metabolize metoprolol. Cultures of 5 strains of Cunninghamella were chosen for this investigation, because such fungi have the ability to metabolize drugs, including verapamil[9], panto-prazole[10], and indomethacin[11], in a similar manner to mammals. The main focus of the current study was to investigate the metabolic fate of metoprolol in cultures of the 5 strains of Cunninghamella and demonstrate parallels of the metabolism of metoprolol in mammalian and in microbial systems.
Materials and methods
Materials Metoprolol (purity 99.0%) was provided by Xinhua Pharmaceutical Co (Changzhou, China); the purity was checked by HPLC analysis. Methanol of HPLC grade was purchased from Concord Technology (Tianjin, China). Peptone and the yeast extract were biochemical reagents. All other chemicals were of analytical grade.
Microorganisms C elegans AS 3.156, C elegans AS 3.2028, C echinulata AS 3.2004, and C blakesleeana AS 3.153 and AS 3.910, were provided by the Institute of Microbiology, Chinese Academy of Sciences (Beijing, China). Stock cultures were maintained on potato dextrose agar (Aoboxing, Beijing, China) slants at 4 °C and transferred every 6 months to maintain viability. The first-stage microbial biotransformation was carried out in a medium consisting of 20 g dextrose, 5 g peptone, 5 g yeast extract, 5 g NaCl, 5 g K2HPO4, and 1000 mL distilled water. The second-stage biotransformation was carried out in a wheat-bran medium containing 1% wheat-bran in broth. The pH of the medium was adjusted to 6.5 with 6.0 mol/L HCl, and then the medium was sterilized in Erlenmeyer flasks (Pierce, Rockford, IL, USA) at 115 °C and 18 psi for 30 min and cooled before incubation.
Biotransformation procedures The microbial metabolism was facilitated by incubating the cultures with shaking on a rotary shaker at 28 °C. For each of the 5 strains of Cunninghamella, the first-stage preculturing was initiated by inoculating a 250-mL Erlenmeyer flask containing 50 mL broth with a loop of spores obtained from a freshly growing agar slant. After incubation for 24 h, a 1.0 mL portion from the first-stage culture was used to inoculate a second-stage 100-mL flask containing 20 mL of broth. The second-stage culture was incubated for 24 h before metoprolol was added to a final concentration of 1.0 g/L. After 120 h of additional incubation, the culture was centrifuged at 1500×g for 20 min, and the supernatant was decanted and kept at -20 °C until analysis.
The preparative-scale biotransformation of metoprolol by C blakesleeana AS 3.153 followed the same procedure as the screening experiments, except for the increase of the broth volume. Two first-stage flasks were prepared as described earlier, and 4 second-stage flasks each containing 100 mL of broth were incubated with 2.5 mL of the first-stage culture. Metoprolol was added to yield a final concentration of 1.0 g/L. The culture was then incubated for an additional 120 h.
Two kinds of controls were conducted simultaneously with the biotransformation procedure. Culture controls consisting of a fermentation blank, in which the microorganisms were grown under the same conditions without metoprolol, were operated to eliminate the interference possibly brought by the microorganism itself or residues of the fermentation cultures. The substrate controls were prepared by adding metoprolol to sterile medium and incubated without the microorganism to determine whether metoprolol could chemically decompose or spontaneously transform under fermentation conditions.
Extraction of metabolites and liquid chromatography-tandem mass spectrometry (LC/MSn) assay A 0.5 mL aliquot of each sample was filtered through a membrane (0.45 µm pore size). The filtrate was applied to a preconditioned Orgchem C18 cartridge (Orgchem, Troy, NY, USA). The cartridge was washed with 1.5 mL water, and the metabolites were eluted with 2.0 mL methanol. A 20 mL aliquot of the eluate was directly injected into the LC/MSn system for analysis.
The LC/MSn analysis was performed on a Thermo Finnigan LCQ ion trap mass spectrometer (San Jose, CA, USA) equipped with an atmospheric-pressure ionization interface. The instrument was operated in positive electrospray ionization (ESI) mode. The spray voltage was set at 4.5 kV. The capillary temperature was maintained at 200 °C and the voltage was fixed at 13 V. The HPLC fluid was nebulized by using N2 as both the sheath gas at a flow rate of 0.75 L/min and the auxiliary gas at a flow rate of 0.15 L/min. A full-scan mass spectrum was operated to obtain the protonated molecules (M+H)+ of each possible metabolite. Multi-stage mass spectra (MS/MS or MS3) were produced by collision-induced dissociation of the selected precursor ions with helium present in the ion trap, and the relative collision energy was set at 30%–35%. Data were collected and analyzed with Xcalibur software (version 1.2, Thermo Finnigan, NJ, USA). Liquid chromatography was carried out with a Shimadzu LC-10AD solvent delivery system (Kyoto, Japan). The samples were separated on a Diamonsil C18 column (200 mm×4.6 mm inner diameter, 5 μm, Dikma, Beijing, China) preceded by a SecurityGuard C18 guard column (4 mm×3.0 mm inner diameter, 5 μm, Phenomenex, Torrance, CA, USA). The mobile phase consisted of methanol-water-formic acid (35:65:0.2, v:v:v) at a flow rate of 0.5 mL/min.
Isolation and identification of major metabolites To isolate the major metabolites of metoprolol in sufficient quantities for structure elucidation, the biotransformation was carried out on a semipreparative scale. After biotrans-formation, the fluid was centrifuged at 1500×g for 20 min. The supernatant was condensed using a Rikakikai Eyela Fdu1100 freeze dryer (Tokyo, Japan). The residue was reconstituted by 10 mL methanol-water (1:1, v:v) and directly deposited onto a 5 cm diameter column packed with Sephadex G10 gel (Zhonghuida Scientific Instrument Co, Dalian, Liaoning, China). The sample was washed exhaustively with methanol and the eluent was collected every 50 mL. The solvent of each fraction was analyzed directly by LC-MSn. Fractions containing possible metabolites were evaporated to dryness under vacuum at 40 °C using a RE-52A rotary evaporator (Yarong Biology Instrument, Shanghai, China). In order to obtain sufficient quantities of metabolites for structural identification, the sample was dissolved in a small amount of water and then injected repeatedly onto a semipreparative HPLC system (Shimadzu, Kyoto, Japan) consisting of 2 LC-6AD solvent delivery units (Shimadzu, Kyoto, Japan), a DGU-14A degasser unit (Shimadzu, Kyoto, Japan), a SCL-10A VP system controller (Shimadzu, Kyoto, Japan), a SPD-10A VP UV-Vis detector (Shimadzu, Kyoto, Japan), a FRC-10A (Shimadzu, Kyoto, Japan) fraction collector, and a CLASS-VP LC workstation (Shimadzu, Kyoto, Japan). Separation was accomplished using a mobile phase consisting of methanol-water (30:70, v:v) at 8.0 mL/min on a Shim-Pack PRC-ODS column (250 mm×20 mm inner diameter, Shimadzu, Kyoto, Japan) preceded by a GPRC-ODS pre-column (8 mm×1.5 mm inner diameter, Shimadzu, Kyoto, Japan). The UV detector was set at 223 nm. The major peaks with similar retention time were pooled, evaporated to dryness, and stored at 4 °C before the structural analysis.
The purified metabolites were dissolved in D2O for the NMR analysis. The NMR measurements were carried out at 600 MHz on a Bruker ARX 600 NMR spectrometer (Bruker, Faellanden, Switzerland). Chemical shifts were reported as parts per million relative to tetramethylsilane as the internal standard.
Results
Screening of cultures Five strains of Cunninghamella were screened and all could transform metoprolol to some extent. The percentages of transformation were 10.1% (C elegans AS 3.156), 12.6% (C elegans AS 3.2028), 82.7% (C blakesleeana AS 3.153), 15.3% (C blakesleeana AS 3.910), and 3.69% (C echinulata AS 3.2004). Since C blakes-leeana AS 3.153 transformed the highest proportion of metoprolol, it was selected for further investigation. The type of medium, pH, and concentration of the substrate were also investigated using an L9 (34) orthogonal table (Table 1). According to Table 1, the first choice for biotransformation should be at pH 4.5 in a wheat-bran medium with a substrate concentration of 1.0 g/L (A2B2C1). However, the factor of pH was found to have no influence on the final yield. Finally biotransformation was carried out at pH 6.5 in a wheat-bran medium with a final substrate concentration of 1.0 g/L.
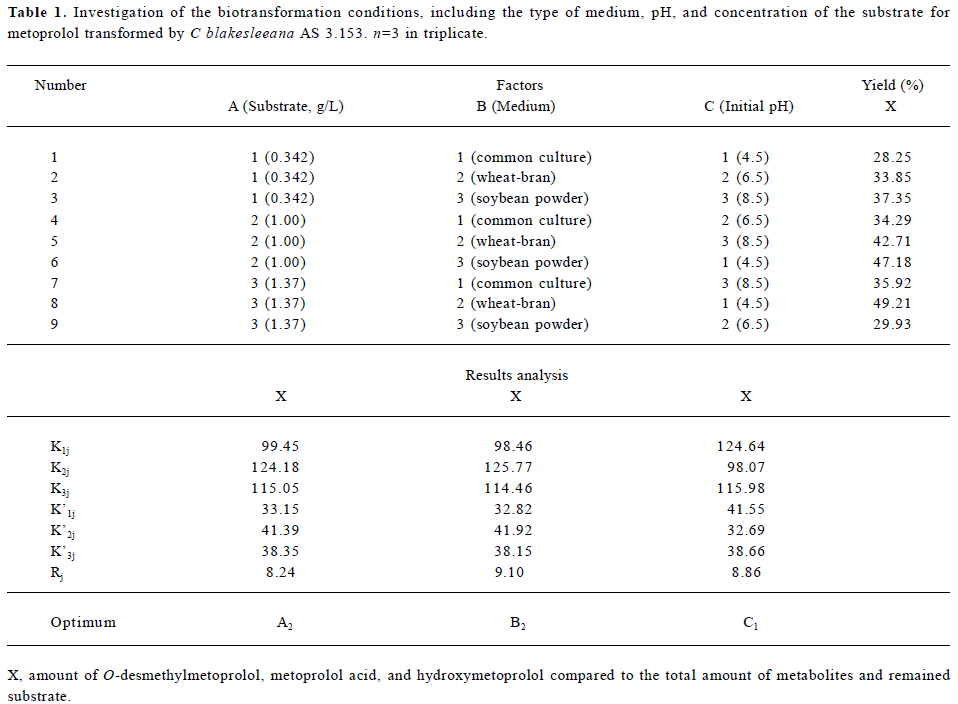
Full table
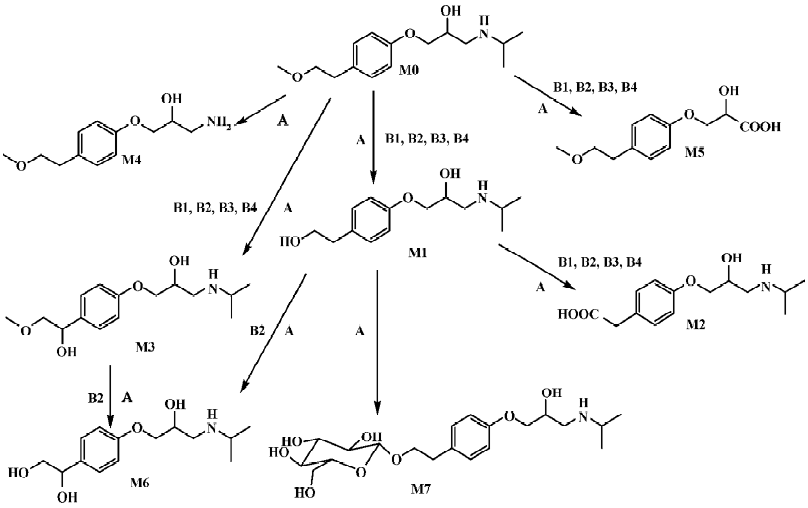
Identification of metoprolol metabolites The LC/MSn chromatograms of the culture control showed no spontaneous formation of possible metabolites of metoprolol under the same conditions. The substrate control contained only metoprolol. As shown in Figure 1, with the exception of metoprolol, 7 possible metabolites were detected in the C blakesleeana AS 3.153 cultures, compared with the control cultures.
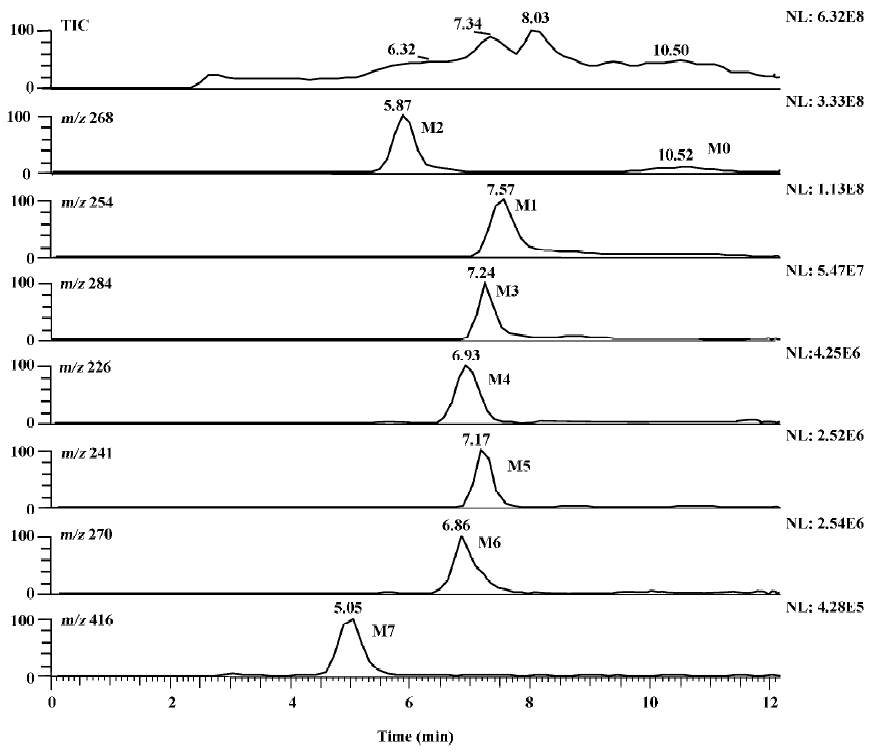
Following the transformation of metoprolol by C blakesleeana AS 3.153, 2 major metabolites (M1 and M2) were isolated by semipreparative HPLC, and their structures were identified by a combination analysis of LC/MSn and NMR spectra. The other metabolites were tentatively identified based on their retention time and MSn information (Table 2).
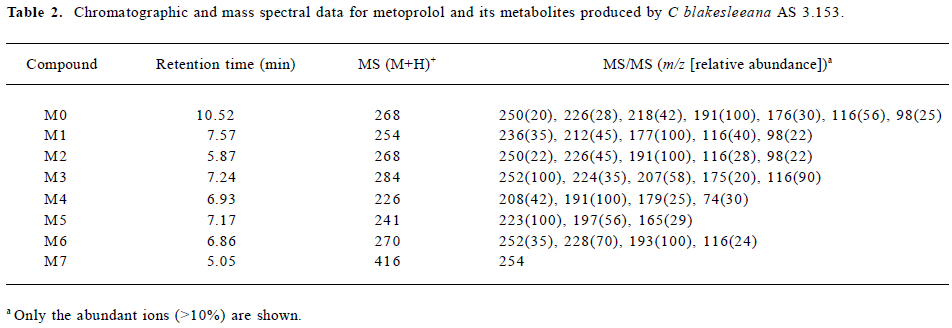
Full table
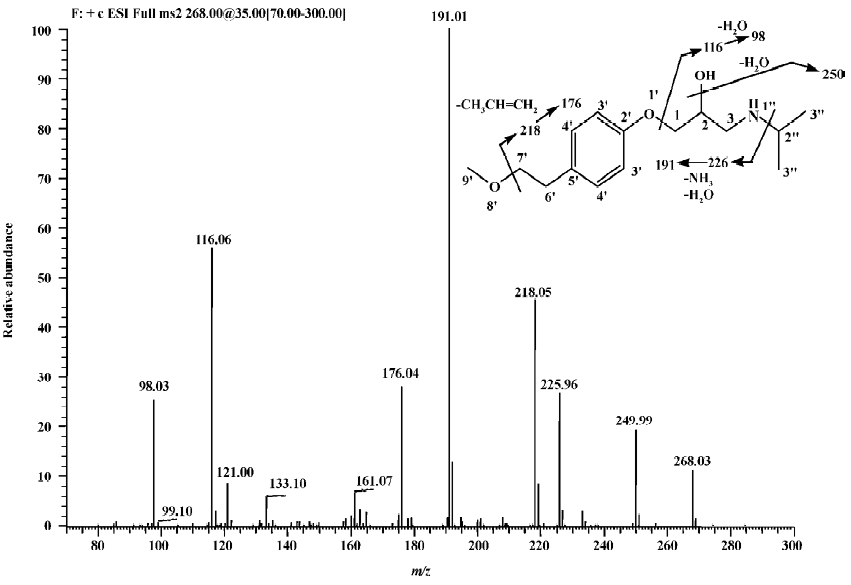
Parent drug The identification of metoprolol (M0) was confirmed by comparison of the retention time and mass spectra (Figure 2) with the authentic reference. The compound eluting at 10.5 min was identified as metoprolol. In order to elucidate the structures of metabolites through the mass spectra, the cleavage pathways of metoprolol were studied first. The MS/MS product ion spectra corresponding to the precursor ion of M0 at m/z 268 and its possible cleavage pathways are shown in Figure 2. The fragment ion at m/z 250 was 18 Da lower than that at m/z 268, which was due to the loss of H2O, and the fragment ion at m/z 218 was formed by further neutral loss of CH3OH, while the fragment ions at m/z 226 and m/z 176 were due to the loss of CH3CH=CH2 according to each precursor ions. The fragment at m/z 191 was due to neutral loss of NH3 and H2O based on the fragment ion at m/z 226. The fragment at m/z 116 was attributed to the elimination of the 4-(2-methoxyethyl)-phenoxy substituent from the structure, whereas the fragment ion at m/z 98 was associated with further loss of H2O.
Metabolite M1 The retention time of M1 ([M+H]+ at m/z 254) obtained by the LC/MSn analysis was 7.57 min. The protonated molecule of M1 was 14 Da lower than that of M0, and its fragment ions at m/z 236, 212, and 177 were also 14 Da lower than that at m/z 250, 226, and 191 of M0, respectively, while the ions at m/z 116 and 98 were the same as that of M0. This suggested that the 1-phenoxy-3-([1-methylethyl]amino)-2-propanol moiety was unchanged. Therefore, M1 was supposed as O-desmethylmetoprolol. To further verify its struc-ture, NMR (1H and 13C) analysis was carried out and the results were as follows: 1H NMR (D2O): δ 0.96 (6H, t, J1=5.28 Hz, J2=4.99 Hz, H-3''), 2.62 (1H, m, H-2''), 2.69 (2H, t, J=6.68 Hz, H-6'), 2.75 (1H, m, H-3), 2.77(1H, m, H-3), 3.67(2H, t, J=6.66 Hz, H-7'), 3.87 (1H, m, H-2), 3.96 (2H, dd, J=5.28 Hz, 18.5 Hz, H-1), 6.85 (2H, d, J=8.30 Hz, H-3'), 7.12(2H, d, J=8.30 Hz, H-4'); 13C NMR (D2O): δ 23.6 (C3''), 23.6 (C3''), 39.6 (C6'), 50.9 (C3), 51.0(C2''), 65.4 (C7'), 71.1 (C1), 73.2(C2), 117.6 (C3'), 117.6 (C3'), 132.9 (C4'), 132.9 (C4'), 134.6 (C5'), and 159.4 (C2'). Compared with the data of metoprolol in previous studies[12], M1 was finally identified as O-desmethyl-metoprolol.
Metabolite M2 The retention time of M2 ([M+H]+ at m/z 268), obtained by the LC/MSn analysis, was 5.87 min. The protonated molecule of M2 was the same as that of M0. The MS/MS spectrum of M2 showed product ions similar to those in M0, except for the absence of m/z 218 and 176, which indicated that changes took place in the methoxyethyl group. To further verify its structure, NMR (1H and 13C) analysis was carried out and the results were as follows: 1H NMR(D2O): δ 1.25 (6H, t, J1=4.99 Hz, J2=5.58 Hz, H-3''), 3.12 (1H, m, H-3), 3.21 (1H, m, H-3), 3.38 (3H, m, H-2'', 6'), 3.98 (1H, dd, J=4.82 Hz, 9.91 Hz, H-1), 4.03 (1H, dd, J=3.73 Hz, 10.1 Hz, H-1), 4.18 (1H, d, J=4.45 Hz, H-2), 6.86 (2H, d, J=8.16 Hz, H-3'), 7.13(2H, d, J=8.14 Hz, H-4'); 13C NMR (D2O): δ 20.6 (C3''), 21.0 (C3''), 46.2(C7'), 49.4(C3), 53.8(C2''), 68.3(C1), 72.2(C2), 117.5 (C3'), 117.5 (C3'), 133.1 (C4'), 133.1 (C4'), 133.2 (C5'), 159.2 (C2'), and 184.0 (C7'). The downfield shift of C7' from 65.4 to 184.0 ppm suggested there was a carboxy group in the structure of M2. Compared with the data of metoprolol in previous studi-es[12], M2 was finally identified as metoprolol acid.
Metabolites M3–M7 The precursor ion of M3 at m/z 284 was 16 Da higher than that of M0, and its fragment at m/z 207 was also 16 Da higher than that at m/z 191 of M0, while the same fragment ion at m/z 116 was in both the MS/MS spectra of M0 and M3, indicating that the 3-([1-methylethyl]amino)-2-propanol moiety was intact, hence, M3 was deduced as α-hydroxymetoprolol. The protonated molecule at m/z 226 (M4) and its fragment ion at m/z 74 were 42 Da lower than that of M0 and its fragment ion at m/z 116, respectively, while there was the same fragment ion at m/z 191, indicating that the metabolism took place at the 3-([1-methylethyl]amino)-2-propanol moiety. Furthermore, the protonated molecule at m/z 226 was identical to the fragment ion at m/z 226 of M0, and MS3 product scan was performed on the fragment ion at m/z 226 of M0. The MS3 spectrum was the same as that of MS2 spectrum of M4, which gave further proof of the structure of M4. Many compounds were found to be N-dealkylated by CYP2D6[13], including other beta-1 selective aryloxy-propano-lamine, such as propranolol[14] and atenolol[15]. N-dealkylation has become an established metabolic pathway, which could also occur on metoprolol. Hence, M4 was tentatively identified as N-desalkylmetoprolol. However, further investigation is needed.
The protonated molecule of M5 was at m/z 241; according to its fragmentation, M5 was supposed to be deaminated metoprolol. To obtain further information, the analysis was operated in negative ESI mode. A peak was detected with the deprotonated molecule at m/z 239. Following these results, M5 was finally identified as deaminated metoprolol. The protonated molecule of M6 at m/z 270 was 16 Da higher than that of M1. The fragment ions of M6 at m/z 252, 228, and 193 were also 16 Da higher than the fragment ions of M1 at m/z 236, 212, and 177, respectively, which suggested that M6 was a hydroxyl metabolite of M1. Similar metabolic pathways also exist in humans, dogs, horses, and rats[2,3].
For M7, the protonated molecule ([M+H]+) was at m/z 416, and 162 Da higher than that of M1. The MS/MS spectra gave only 1 major fragment at m/z 254, which may be due to the neutral loss of glucose. Therefore, the MS3 product scan was performed on m/z 254, and 3 major fragment ions at m/z 236, 212, and 177 were observed (data not shown), which was similar to the MS/MS spectra of M1. The retention time of M7 (5.05 min) was ahead of other metabolites, which was possibly due to its increasing polarity. These data suggested that M7 was the glucoside conjugate of M1. The eluate of M7 was collected and evaporated to dryness under a gentle stream of nitrogen. The residue was incubated with β-D-glucosidase (100 kU/L) at 37 °C for 24 h. The incubation solution was analyzed by LC/MSn. The peak of M7 disappeared; instead, a peak with the same MS spectra and retention time as that of M1 was detected, which provided further evidence that M7 was the metabolite conjugated with glucoside.
Microbial transformation of metoprolol and comparison with mammalian metabolism In the present study, metoprolol was transformed by C blakesleeana AS 3.153 to 7 metabolites: O-desmethylmetoprolol (M1), meto-prolol acid (M2), α-hydroxymetoprolol (M3), N-desalkyl-metoprolol (M4), deaminated metoprolol (M5), hydroxyl-O-desmethylmetoprolol (M6), and glucoside conjugate of O-desmethylmetoprolol (M7). As shown in Figure 3, the structures of the metabolites and proposed biotransformation pathways by C blakesleeana are compared to those that have been identified in mammals[2,3]. After 120-h incubation by C blakesleeana, about 80% metoprolol was metabolized mainly to 3 metabolites, which was consistent with that in mammals. The yields of these metabolites were M1 (21.2%), M2 (59.4%), and M3 (4.5%), respectively.
Discussion
We reported a successful biotransformation of metoprolol by C blakesleeana AS 3.153 in this study. After centrifu-gation, the layer of mycelium was analyzed and trace amounts of metoprolol and its metabolites (M1 and M2) were detected. About 96% of the substrate and metabolites were obtained in the supernatant after centrifugation according to the initial amount of metoprolol. Thus, the analysis was performed only in the supernatant. After 120-h incubation in wheat-bran broth, about 80% of the drug was metabolized to 7 metabolites. As shown in Figure 3, 5 of the metabolites were essentially similar to those obtained in mammalian metabolism studies, whereas 2 novel metabolites, N-desalkyl-metoprolol and the glucoside conjugate of O-desmethyl-metoprolol, were identified. It has been well recognized that conjugation is an important metabolic pathway of many compounds both in mammals and in microorganisms[16–18]. A glucoside conjugate was detected in the present study, which conformed to previous studies that the glucosidation of drugs can be formed by microbial models[10,18,19]. In humans, metoprolol was metabolized to O-desmethylmeto-prolol and metoprolol acid or α-hydroxymetoprolol, depending on the cytochrome P450 oxidation phenotype. The 3 primary metabolites of metoprolol in rats, dogs, and horses were O-desmethyl-metoprolol, metoprolol acid, and α-hydroxymetoprolol[2,3]. The fungus C blakesleeana converted metoprolol to 3 major metabolites, which showed similarities with the metabolism of metoprolol in mammals.
In conclusion, 7 metabolites of metoprolol were formed by C blakesleeana AS 3.153. The ability of C blakesleeana to mimic the mammalian metabolism and to perform novel biotransformations clearly demonstrated that microbial systems could predict potential routes of the mammalian biotransformation of drug candidates, and could also be used for the understanding of chemistry and biology significance of drug metabolism. Because of the capacity of the microbial metabolism, substrate concentrations used are much higher than those employed in other cell or tissue models and consequently allow for easier detection and isolation. Thus, the models can be scaled up easily for the preparation of metabolites for structure confirmation by NMR and further pharmacological and toxicological studies.
Acknowledgement
We thank Ms Yu-ya WANG for her assistance on the interpretation of the NMR spectrum.
References
- Davis ME, Richards AM, Nicholls MG, Yandle TG, Frampton CM, Troughton RW. Introduction of metoprolol increases plasma B-type cardiac natriuretic peptides in mild, stable heart failure. Circulation 2006;113:977-85.
- Borg KO, Carlsson E, Hoffmann KJ, Jonsson TE, Thorin H, Wallin B. Metabolism of metoprolol (3H) in man, the dog and the rat. Acta Pharmacol Toxicol 1975;36:125-35.
- Dumasia MC. In vivo biotransformation of metoprolol in horse and on-column esterification of the aminocarboxylic acid metabolite by alcohols during solid phase extraction using mixed mode columns. J Pharm Biomed Anal 2006;40:75-81.
- Lennard MS, Tucker GT, Woods HF. The polymorphic oxidation of beta-adrenoceptor antagonists. Clinical pharmacokinetic considerations. Clin Pharmacokinet 1986;11:1-17.
- Lisowska K, Szemraj J, Rozalska S, Dlugonski J. The expression of cytochrome P-450 and cytochrome P-450 reductase genes in the simultaneous transformation of corticosteroids and phenanthrene by Cunninghamella elegans. FEMS Microbiol Lett 2006;261:175-80.
- Zhang D, Zhang H, Aranibar N, Hanson R, Huang Y, Cheng PT, et al. Structural elucidation of human oxidative metabolites of muragli-tazar: use of microbial bioreactors in the biosynthesis of metabolite standards. Drug Metab Dispos 2006;34:267-80.
- Lisowska K, Dlugonski J, Freeman JP, Cerniglia CE. The effect of the corticosteroid hormone cortexolone on the metabolites produced during phenanthrene biotransformation in Cunning-hamella elegans. Chemosphere 2006;64:1499-506.
- Chen Y, Monshouwer M, Fitch WL. Analytical tools and appro-aches for metabolite identification in early drug discovery. Pharm Res 2007;24:248-57.
- Sun L, Huang HH, Liu L, Zhong DF. Transformation of verapa-mil by Cunninghamella blakesleana. Appl Environ Microbial 2004;70:2722-7.
- Xie ZY, Huang HH, Zhong DF. Biotransformation of panto-prazole by the fungus Cunninghamella blakesleana. Xenobiotica 2005;35:467-77.
- Zhang P, Lin LH, Huang HH, Xu HY, Zhong DF. Biotransformation of indomethacin by the fungus Cunninghamella blakes-leeana. Acta Pharmacol Sin 2006;27:1097-102.
- Kulkarni VM, Coutinho EC. Conformational analysis of atenolol and metoprolol by 1H NMR. Indian J Chem 1991;30:52-6.
- de Groot MJ, Ackland MJ, Horne VA, Alex AA, Jones BC. A novel approach to predicting P450-mediated drug metabolism. CYP2D6 catalyzed N-dealkylation reactions and qualitative metabolite predictions using a combined protein and pharmacophore model for CYP2D6. J Med Chem 1999;42:4062-70.
- Upthagrove AL, Nelson WL. Importance of amine pKa and distribution coefficient in the metabolism of fluorinated propranolol derivatives. Preparation, identification of metabolite regio-isomers, and metabolism by CYP2D6. Drug Metab Dispos 2001;29:1377-88.
- Bonde J, Bodtker S, Angelo HR, Svendsen TL, Kampmann JP. Atenolol inhibits the elimination of disopyramide. Eur J Clin Pharmacol 1985;28:41-3.
- Zamek-Gliszczynski MJ, Hoffmaster KA, Nezasa K, Tallman MN, Brouwer KL. Integration of hepatic drug transporters and phase II metabolizing enzymes: mechanisms of hepatic excretion of sulfate, glucuronide, and glutathione metabolites. Eur J Pharm Sci 2006;27:447-86.
- Levsen K, Schiebel HM, Behnke B, Dotzer R, Dreher W, Elend M, et al. Structure elucidation of phase II metabolites by tandem mass spectrometry: an overview. J Chromatogr A 2005;1067:55-72.
- Huang HH, Ma GL, Sun YM, Li Q, Zhong DF. Phase II metabolites of etofesalamide in filamentous fungi. Acta Pharmacol Sin 2005;26:893-6.
- Zhan J, Gunatilaka AA. Microbial transformation of curvularin. J Nat Prod 2005;68:1271-3.
