Effect of Porphyromonas gingivalis PrtC on cytokine expression in ECV304 endothelial cells and its level in subgingival plaques from patients with chronic periodontitis1
Introduction
A significant percentage of the human population suffers from periodontitis, a chronic inflammatory disease characterized by the breakdown of periodontal tissue[1]. The incidence and rate of progression of this disease involve complex interactions among periodontopathic bacteria as well as between periodontopathic bacteria and host cells[2,3]. These interactions lead host cells to release a broad array of inflammatory cytokines, chemokines, and mediators, some of which result in the destruction of periodontal supporting tissue, namely alveolar bone, periodontal ligament, and cementum around teeth[2,3]. The Gram-negative anaerobic bacterium Porphyromonas gingivalis is implicated as one of the important etiological agents of chronic and aggressive forms of periodontitis[4–7]. This organism expresses a number of potential virulence factors, including fimbriae, superoxide dismutase, hemagglutinin, the arginine-X-specific cysteine proteases, and the lysine-X-specific proteases[8]. In addition, collagenolytic activity of P gingivalis has been described and is associated with the production of several proteases[8]. Since collagen is an important component of the periodontium, collagenase activity plays a critical role in tissue destruction and progression of periodontitis[9]. Specific cleavage of type I collagen has been attributed to the function of the prtC gene product, which is referred to as collagenase[8]. Takahashi and Kato were the first to report the cloning and expression of the prtC gene from P gingivalis ATCC 33277[10,11]. While the prtC gene product did not exhibit structural similarity with eukaryotic collagenases, it was able to degrade soluble and reconstituted fibrillar type I collagen, heat-denatured type I collagen, and azocoll, but did not degrade synthetic collagenase substrates and did not contain a partial zinc-binding region that is consensus in these enzymes[12]. These features may reflect the unique nature of this collagenase or perhaps question its status as a true collagenase[12]. Nonetheless, the inability of a collagenolytic enzyme to degrade native fibrillar collagen does not preclude its involvement in the pathogenesis of periodontal disease[12]. Proteinase produced by some bacteria could have a direct effect on inducing inflammation besides its specific enzyme activity[13] and may work in combination with true bacterial or host collagenases[14,15]. Recent findings indicate that P gingivalis induces host collagen degradation by affecting expression, activation, and inhibition of matrix metalloproteinases (MMP) produced by host cells[8,16]. MMP is thought to be transcriptionally upregulated by pro-inflammatory mediators, such as interleukin (IL)-1 and TNF-α, as well as post-translationally activated by proteases from P gingivalis[16]. However, studies on the prtC gene product of P gingivalis mainly concentrate on its collagenolytic activity. Its ability to induce host cells to produce inflammatory cytokines has yet not been examined, which is of great importance in understanding its role in the pathogenesis and development of periodontitis.
Several studies have demonstrated that P gingivalis is able to invade and activate different cell types in the surrounding tissue of teeth, including endothelial, gingival epithelial cells, as well as periodontal ligament cells[14,15,17]. Endothelial cells, therefore, can act as primary target cells during infection with P gingivalis. Endothelial cells are key players during inflammatory reactions and are able to produce an array of pro-inflammatory mediators, including cytokines such as IL-1, IL-6, IL-8, TNF-α, and lipid mediators like prostaglandins or platelet-activated factor[17,18]. A previous report has indicated that P gingivalis can infect human umbilical vein endothelial cells (HUVEC) and trigger a cascade of events that could lead to endothelial damage, as well as local and systemic inflammation[17]. Moreover, host cells sense live P gingivalis and its components such as lipopolysaccharides (LPS) or fimbriae differently, which translate into the expression of different inflammatory cytokine profiles[19]. The objective of the present work is to investigate the effect of the P gingivalis PrtC protein on the expression of cytokines in HUVEC-originated ECV304 cells, which has been shown to be a suitable in vitro model for the study of endothelial cells[20,21,22], and subsequently, to determine the possible association of P gingivalis PrtC levels with periodontal conditions.
Materials and methods
In vitro study
P gingivalis strains and growth condition The P gingivalis strain ATCC 33277 was supplied by the Department of Medical Microbiology and Parasitology, College of Medicine, Zhejiang University, China. They were inoculated on trypticase soy agar supplemented with 5 µg/mL haemin, 1 µg/mL vitamin K1, 5% (v/v) sheep blood, and 1 µg/mL menadione. The plates were incubated in an anaerobic chamber under atmosphere condition of 80% N2, 10% CO2, and 10% H2 at 37 °C for 7 d.
Cloning, sequencing and construction of recombinant expression vector The P gingivalis prtC was amplified from type strain ATCC 33277 by PCR with a forward primer containing the site of BamH I: 5'-GGG
The prtC fragment was cloned into the pUCm-T vector by using the T-A cloning kit according to the manufacturer’s instructions (Biocolor, Shanghai, China). The recombinant plasmid (pUCm-T-prtC) was first transformed into the Escherichia coli strain DH5α. After nucleotide sequencing confirmed the inserted fragments, the E coli DH5α strains containing pUCm-T-prtC or the prokaryotic expression vector pET32a were amplified and then the plasmids were extracted. pUCm-T-prtC and the pET32a vector were both digested with BamHI and XhoI. The target fragments of the prtC gene and pET32a were recovered and then ligated with a ligation kit (TaKaRa Biomedicals, Otsu, Shiga, Japan). The recombinant expression vector pET32a-prtC was transformed into E coli strain BL21 (DE3). Then the plasmids were extracted and sequenced again.
Expression and identification of the target recombinant protein The constructed prokaryotic expression system pET32a-prtC-E coli BL21DE3 was rotationally cultured in Luria-Bertani (LB) medium at 30 °C under inducement of 0.5 mmol/L isopropyl-1-thio-β-galactoside (IPTG). The supernatant and precipitate were isolated by centrifugation after the bacterium was ultrasonically broken. SDS-PAGE (10%) was used to examine molecular weight, output, and location of the target recombinant protein (rPrtC). rPrtC was then collected and purified by affinity chromatography on a Ni-NTA column (QIAGEN, Hilden, Germany). Following elution with imidazole, contaminated LPS was removed by Triton X-114 extraction. Then the protein was chromatographed on Sephadex G25 columns (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and eluted in phosphate-buffered solution. Solution of the recombinant protein 10 µg/mL was found to be free of endotoxins by the Limulus amoebocyte lysate assay (Bio Whittaker, Walkersville, MD, USA). Rabbits were routinely immunized with 1 mg purified rPrtC or 1 mg whole cells of P gingivalis supplemented with complete Freund’s adjuvant, administered subcutaneously 4 times. Slide agglutination reaction and ELISA were used to detect the immunoreactivity of rabbit anti-rPrtC antiserum against P gingivalis ATCC 33277, Prevotella melaninogenica ATCC 25845, Fusobacterium nucleatum ATCC 25586, E coli ATCC 25922, Actinomycetemcomitans actinobacillus Y3, Veillonella parvula 990116, and 100 µg/mL of their sonicated supernatants, respectively. After electrophoresis on 10% SDS-PAGE, the rPrtC protein was then transferred to a polyvinylidene fluoride (PVDF) membrane using the Trans-Blot SD apparatus (Bio-Rad, Richmond, CA, USA). The blot was blocked for 1 h in Tris-buffered saline containing 0.1% Tween-20 (TBST) and 5% (w/v) skim milk and then incubated with the rabbit antisera (1:1500 dilution) for 1 h. After washing unbound primary antibodies with TBST 3 times for 10 min each, the blot was treated with horseradish peroxidase (HRP)-conjugated sheep anti-rabbit immunoglobin G (1:3000 dilution; Jackson ImmunoResearch, West Grove, PA, USA) and developed with the ECL system (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
ELISA detection of inflammatory cytokine secretion in ECV304 cells induced by rPrtC HUVEC ECV304 was maintained at a density of 5×103 cells/well in a 96-well plate in RPMI-1640 medium (Gibco BRL, Gaithersburg, MD, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 in air at 37 °C for 24 h. Then the ECV304 cells were cultured in RPMI-1640 medium supplemented with 2% FBS which contained 1, 5, and 10 µg/mL rPrtC for 12, 24, or 48 h, respectively. Each concentration and time interval was repeated 3 times. The supernatants were collected for the ELISA detection of IL-1α, IL-8, and TNF-α (R&D Systems, Minneapolis, MN, USA). Recombinant TNF-α (0.1µg, SibEnzyme, Novosibirsk, Siberia, Russia) and RPMI-1640 medium containing 2% FBS were used as positive and negative controls, respectively.
In vivo study
Patients The patients were 49 Chinese chronic periodontitis (CP) patients [24 males aged 29–66 years (mean age, 45.7±8.9 years) and 25 females aged 33–67 years (mean age, 44.3±7.6 years)], and 25 individuals with healthy periodontium [11 males aged 26–64 years (mean age, 41.8±5.8 years) and 14 females aged 22–63 years (mean age, 41.3±6.8 years)] who were referred to the dental clinic in the Second Affiliated Hospital, College of Medicine, Zhejiang University for dental or periodontal treatment or health monitoring. All the patients were non-smokers without any systemic disease, and had at least 14 teeth. Those who had received professional cleaning or had history of antibiotic therapy during the preceding 3 months were excluded. All of the patients and the healthy individuals underwent a full mouth examination. The criteria of diagnosis for chronic periodontitis were based on the Classification of the Periodontal Diseases issued by American Academy of Periodontology in 1999[23]. Briefly, the 49 generalized CP patients had >30% sites showing periodontal probing depth ≥3 mm, clinical attachment loss >1 mm, and radiographic evidence of alveolar bone loss. Individuals with periodontal probing depth less than 3 mm without any clinical attachment loss or radiographic evidence of bone loss and without inflammation of gingivae were considered healthy. All the patients received detailed information concerning the nature of the study and the procedures involved, and their informed consent was obtained. The ethical committee of the College of Medicine at Zhejiang University approved the study protocol.
Collection of samples For each patient, 4 subgingival plaque samples were taken from the bottom of the 4 deepest periodontal pockets of the dentition, preferably 1 pocket from each quadrant, with separate sterile Gracy curettes after supragingival plaque was gently removed. After initial periodontal treatment that mainly included oral hygiene instruction, full mouth supragingival and subgingival scaling and root planning, subgingival plaque samples from the same sites were collected again 4 months later. For individuals with healthy periodontium, samples from the bottom of gingival sulcular were taken with the same method. Each of the samples was placed in 200 µL lysis buffer, which consisted of 1 mmol/L EDTA, 1% Triton X-100, and 10 mmol/L Tris-HCl (pH 8.0), and stored at -20 °C until use. Clinical parameters, such as probing depth (PD), attachment loss (AL), and bleeding on probing (BOP) at 6 sites of each tooth of the full dentition were recorded at baseline and after initial periodontal treatment. The 6 sites were the mesial-buccal, middle-buccal, distal-buccal, mesial-lingual/palatal, middle-lingual/palatal, and distal-lingual/palatal sites of each tooth.
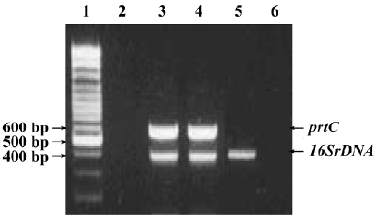
PCR detection of P gingivalis in subgingival plaque samples Lysis buffer 100 µL containing subgingival plaque samples was boiled for 10 min, and 10 µL supernatant was directly used as a template in the PCR. A multiplex PCR assay was established to detect the P gingivalis 16SrDNA and prtC genes. The sequences of the primers specific for the P gingivalis 16SrDNA gene were: 5'-AGG CAG CTT GCC ATA CTG CG-3' (sense) and 5'-ACT GTT AGC AAC TAC CGA TGT-3' (antisense)[24]. For the prtC gene they were: 5'-ACA ATC CAC GAG ACC ATC-3' (sense) and 5'-TTC AGC CAC ACC GAG ACG-3' (antisense)[25]. PCR amplification was performed in a total volume of 100 µL, containing 10 µL of the template, 10 µL PCR buffer (pH 8.3), and 3U EX-Taq polymerase (TaKaRa Biomedicals, Japan), 0.25 mmol/L dNTP, 2.5 mmol/L MgCl2, 250 nmol/L primers each for P gingivalis 16SrDNA and prtC genes. The PCR programs for the detection of P gingivalis included an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, primer annealing at 52 °C for 30 s, extension at 72 °C for 1 min, and then a final extension step at 72 °C for 7 min. In each of the PCR assays, 10 ng P gingivalis ATCC 33277 DNA preparation was co-amplified with the subgingival plaque samples as the positive control for the detection of P gingivalis. The expected sizes of the PCR products amplified from the P gingivalis 16SrDNA and prtC genes was 404 bp and 584 bp[24,25]. Each reaction product 10 µL were mixed with 10 µL of 2×loading buffer and fractionated in a 2% agarose gel containing ethidium bromide (1 µg/mL), using a 100 bp DNA ladder (Promega, Madison, WI, USA) as a size marker, and visualized under UV light.
ELISA detection of PrtC levels in subgingival plaque samples The subgingival plaque samples were sonicated, and the protein concentration in each of the samples was measured by UV spectrophotometry. Each sample was diluted to 20 µg/mL protein with coating buffer in ELISA. By using 0.1 mL of the diluted samples as the coating antigen, the self-prepared rabbit anti-rPrtC serum (1:1000 dilution) as the first antibody, and HRP-labeled sheep anti-rabbit IgG (1:3000 dilution) as the second antibody, the PrtC levels in the samples were detected. A model 680 microplate reader (Bio-Rad, USA) was used to detect the A490 value. The ELISA result of a sample was considered positive if its A490 value was over the mean plus 3 times that of standard deviation (SD) of the sonicated sulcular samples with the same protein concentration from the 25 individuals with healthy periodontium who were used as negative controls in the test. Five wells of 20 µg/mL rPrtC were used as the positive control.
Statistical analysis The clinical and laboratory data were presented as mean±SD. Student’s t-test and ANOVA were used to determine the significance of the differences between the sub-groups. P<0.05 was considered statistically significant.
Results
PCR detection of P gingivalis in subgingival plaque samples The 25 individuals with healthy periodontium were all negative for P gingivalis. In the 196 subgingival samples from CP patients, 95.9% (188/196) and 91.8% (180/196) of the samples were positive for P gingivalis 16SrDNA and prtC, respectively. None of the samples were positive for prtC and negative for 16SrDNA (Figure 1).
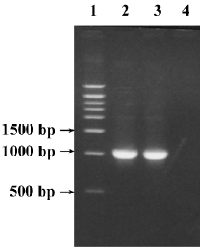
Nucleotide sequence analysis The target fragments with the expected size amplified from genomic DNA of P gingivalis strain ATCC 33277 are shown in Figure 2. Compared with the published sequences (GenBank Accession N
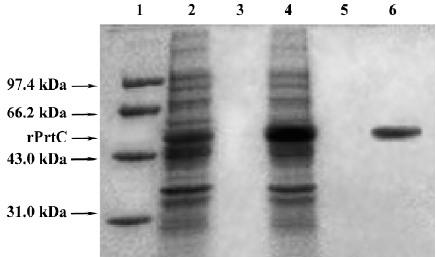
Expression of target fusion protein IPTG 0.5 mmol/L was able to efficiently induce rPrtC expression (Figure 3). Purified rPrtC showed 1 single band on 10% SDS-PAGE. rPrtC was mainly presented in ultrasonic precipitation and the output was approximately 50% of the total bacterial proteins.
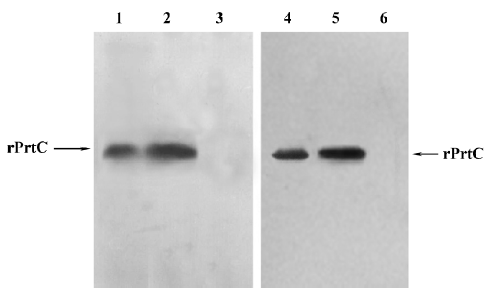
Antigenicity and immunoreactivity of rPrtC The rabbit anti-rPrtC antisera was found positive for P gingivalis ATCC 33277 by slide agglutination reaction; the A490 value of the supernatants was 0.89±0.10 as determined by ELISA. The slide agglutination reactions with the other 5 species were all negative; the A490 value was 0.01~0.03±0.02~0.03, which indicated that the antisera were specific. Furthermore, Western blotting indicated that rPrtC could combine with the rabbit anti-rPrtC serum and rabbit antiserum against the whole cell of P gingivalis, and only one distinct band was shown(Figure 4). It demonstrated that rPrtC could induce rabbit to produce specific antiserum, showing good antigenicity of the recombinant protein. The fact rPrtC could combine with these antiserum also implied its immunoreactivity.
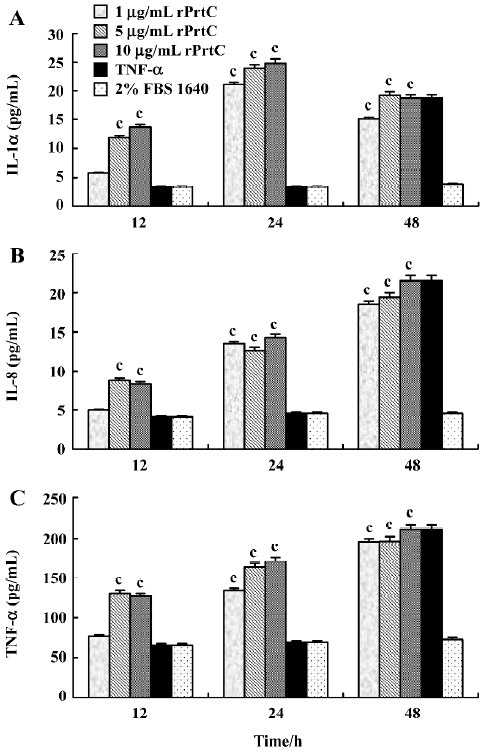
Effects of rPrtC on the secretion of inflammatory cytokines in ECV304 cells After the ECV304 cells were treated with 1 µg/mL rPrtC for 24 h or with 5 and 10 µg/mL rPrtC for 12 h, the secretion of IL-1α, IL-8 and TNF-α in the supernatants increased significantly (P>0.05), among which the IL-1α level peaked at 24 h and the IL-8 and TNF-α levels increased gradually with time. Slightly higher levels of IL-1α and TNF-α were shown in the ECV304 cells stimulated with 5 or 10 µg/mL rPrtC than with 1 µg/mL rPrtC for the same time intervals (Figure 5).
PrtC level in subgingival plaque samples before and after treatment The mean±SD at A490 of the negative sulcular samples was 0.06±0.06, and the positive reference value was 0.24. According to this, 90.8% (178/196) of the subgingival plaque samples were positive for PrtC with an A490 value ranging from 0.26 to 1.23. All these 178 PrtC positive results were from prtC gene positive samples. Only 2 samples were PrtC negative, but were prtC gene positive. The correlation between the levels of P gingivalis PrtC in subgingival plaques and clinical parameters are shown in Table 1. The A490 value in the BOP-positive sites was significantly higher than that in BOP-negative sites (P=0.029). The enhanced level of PrtC was found in the ≥5 mm AL sites than that in the ≤2 mm AL sites (P=0.016), but with no significant difference between the ≥5 and >2–<5 mm AL sites or between the ≤2 and >2–<5 mm AL sites (P=0.114, 0.362). A higher A490 value was detected in the >6 mm PD pockets than in >4–≤6 mm or in the 3–4 mm sites (P=0.039, 0.023). No statistical difference could be distinguished between the >4–≤6 mm and the 3–4 mm pockets (P=0.506).
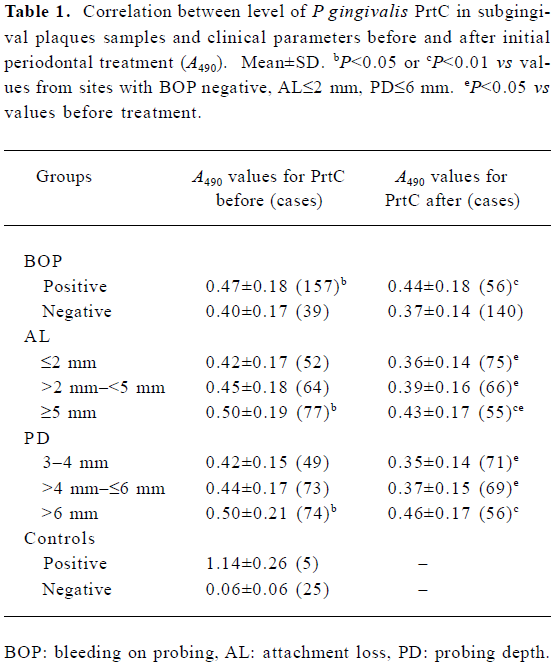
Full table
Overall, the periodontal conditions in the periodontitis patients were improved significantly 4 months after the initial periodontal treatment (Table 2). Only 56 of the 157 sites were still positive for BOP (Table 1). The A490 value decreased after treatment, with 66.8% (131/196) of the samples positive for PrtC in a range of 0.25–0.88. Although the PrtC level in the BOP-positive sites was still higher than that in the BOP-negative sites after treatment (P=0.004), no significant difference was found between the A490 values before and after treatment (P=0.261, 0.286). Higher values were detected in the ≥5 mm AL sites than those in ≤2 mm AL sites (P=0.011), but with no statistical difference between the ≥5 and the >2–<5 mm AL sites or between the ≤2 and the >2–<5 mm AL sites (P=0.186, 0.237). A remarkably enhanced level of PrtC was observed in the >6 mm PD pockets than that in the >4–≤6 mm or in the 3–4 mm pockets (P=0.002, 0.000), but the A490 values between the >4–≤6 mm and the 3–4 mm PD pockets were almost similar (P=0.416). After treatment, the PrtC level in the different AL groups or the ≤6 mm PD groups decreased markedly compared with that before treatment (P=0.010–0.047). Although the A490 values also dropped slightly compared with that before treatment in the sites with >6 mm PD, no statistical difference could be found (P=0.246).
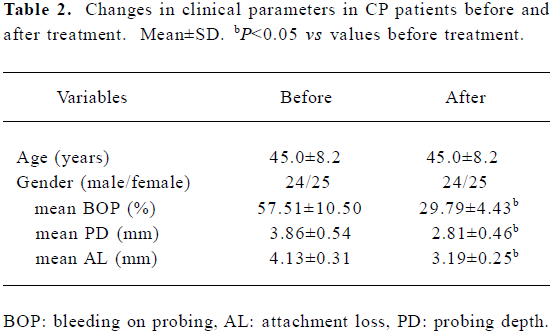
Full table
Discussion
In the present study, a prokaryotic expression system was constructed to obtain plentiful purified recombinant PrtC, which would be helpful in investigating the potential role of P gingivalis collagenase in periodontal destruction, because it was difficult to extract a great deal of purified collagenase directly from P gingivalis. A nucleotide sequence analysis showed 98.46% and 99.07% homology of the nucleotide and putative amino acid sequences of our cloned prtC gene compared with what was registered in GenBank, indicating high fidelity of the method. A low dosage of 0.5 mmol/L IPTG could efficiently induce rPrtC expression with a 50% output of the bacterial total proteins, suggesting the efficiency of the constructed prokaryotic expression system. The fact that rPrtC could be recognized by the antibody against the whole cell of P gingivalis and that it was able to induce rabbit antisera to produce the specific antibody, demonstrated good immunoreactivity and antigenicity of the recom-binant protein. A high frequency of infection of P gingivalis in the subgingival plaque sample in CP patients (95.9%) was revealed by PCR. In the 196 samples, 188 and 180 were positive for P gingivalis 16SrDNA and prtC, respectively. The data implied that the prtC gene might be present in most P gingivalis strains (180/188), which was consistent with previous reports[25,26]. Furthermore, compared with the PCR detection of the prtC gene, our established ELISA was highly sensitive. Among the 180 samples positive for the prtC gene, 178 were found to be PrtC positive by ELISA. This indicated that the prtC gene was frequently expressed in this species.
This is the first study to demonstrate that rPrtC is able to directly promote the production of IL-1α, IL-8 and TNF-α in endothelial cells. Initial studies indicated that the prtC gene product could cleave type I collagen, suggesting that this enzyme might play a role in connective tissue destruction in periodontitis[4,11]. More recent findings however questioned the role of the prtC gene product in the collagenolytic activity of P gingivalis[27,28]. Some researchers have found that proteinase from some bacteria could have direct inflammation-causing ability on host cells besides its specific enzyme activity[13,29]. However, studies of the influence of the prtC gene product of P gingivalis on host cells are still lacking. On the other hand, neutrophil-endothelial cell interactions are the prerequisite for the transendothelial migration of leukocytes, activation of T cells, and then establishment of local inflammation[29], so endothelial cells play a critical role in the development of inflammation. Moreover, endothelial cells may be important target cells for periodontal pathogens, including P gingivalis[17]. In this study, we investigated the effect of recombinant PrtC on the secretion of cytokines in ECV304 endothelial cells. After co-incubation with 1 µg/mL rPrtCfor 24 h and with 5 or 10 µg/mL rPrtC for 12 h, the levels of IL-1α, IL-8, and TNF-α increased significantly (P<0.05). IL-1α and TNF-α exert multiple effects on inflammatory cells, including the synthesis of other cytokines and the modulation of intracellular adhesion molecule-1 upregulation, a critical step in the development of neutrophil-endothelial cell interactions[13,19,29]. IL-8 is key mediator of neutrophil migration to sites of inflammation[13,19]. IL-8 is produced in response to cytokines such as TNF-α, IL-1, and LPS[13,19]. So the ability of PrtC to induce the expression of these cytokines in endothelial cells implies an important role of PrtC in the inflammation of periodontal tissue. Much attention has been given to proteinase produced by host cells. MMP, enzymes secreted by endothelial cells, macrophages, and fibroblasts, are known to be involved in tissue remodeling, organo-genesis, angiogenesis, wound repair, and inflammatory cellular infiltration by degrading extracellular matrix[16,30]. At the inflammatory site, inflammatory cells are mobilized and MMP are produced as pro-enzymes and activated by various stimulatory factors[30]. Recent studies demonstrated that MMP expression was upregulated by pro-inflammatory mediators including IL-1 and TNF-α[16]. Taken together, these findings suggest that although the prtC gene product may not be a true bacterial collagenase, it could have a strong effect on inducing host cells to secret inflammatory cytokines and further enhance the collagenase activity of host cells such as MMP, which lead to the destruction of periodontal connective tissue.
Our data further demonstrated that the PrtC level in clinical samples was correlated with BOP, AL, and PD before and after treatment (Table 1). ELISA, with the self-prepared rabbit anti-rPrtC serum, was established to measure the PrtC level in subgingival samples to understand the association between PrtC and periodontal inflammation or tissue destruction. The PrtC level was higher in the BOP-positive sites or in serious periodontal destruction sites (≥5 mm AL) than in the BOP-negative or in the ≤2 mm AL sites (P<0.05). The A490 value of PrtC was higher in the deep pockets (>6 mm PD) than that in shallow or moderate pockets (P<0.05). This indicates that the PrtC level is related to periodontal inflammation and tissue destruction. A previous study in domestic cats showed that serum antibody response to the recombinant PrtC of a feline strain of P gingivalis was associated with the severity of periodontal disease[12]. The present in vivo study provided strong evidence that PrtC correlated with periodontal tissue destruction and inflammation. PrtC might play a role in the initiation and development of periodontitis by working in combination with other factors produced by P gingivalis or host cells, including P gingivalis LPS or MMP[31,32], since the relationships between PrtC levels and these clinical parameters seem to be rather com-plicated. It was notable that the PrtC-positive subgingival samples were reduced from 90.8% to 66.8%, and the PrtC level decreased remarkably in different AL sites or in the ≤6 mm PD pockets (P<0.05) after treatment, which might be explained by the fact that mechanical debridement will significantly decrease the number of P gingivalis and other putative periodontal pathogens[32,33]. However, the A490 value of PrtC in the BOP-positive samples and in the sites with >6 mm PD changed insignificantly (P>0.05). Previous studies have indicated that deep pockets and furcations are most likely inadequately instrumented[34] and microorganisms, including P gingivalis residing in biofilms left in such locations, correlated with BOP and greater pocket depths[35], which may require surgical intervention[32]. Our results also contributed to the above viewpoints.
In conclusion, periodontal diseases are multifactorial infections elicited by a complex of bacterial species that interact with host tissues and cells, which lead to secretion of various cytokines, including IL-1, IL-6, IL-8, and TNF-α, as well as prostaglandin E2 and MMP[2,3,8]. These cytokines and enzymes play critical roles in periodontal tissue destruc-tion. The results obtained in this study demonstrated that the prtC gene product could induce host cells to synthesize and secrete inflammatory cytokines to exert a destructive effect on periodontal tissue. Further investigation is needed to reach a conclusive result.
Acknowledgement
We would like to thank Dr Wei-lian SUN for assistance in the recruitment of patients and the collection of clinical samples.
References
- Kumagai Y, Yagishita H, Yajima A, Okamoto T, Konishi K. Molecular mechanism for connective tissue destruction by dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis. Infect Immun 2005;73:2655-64.
- Socransky SS, Haffajee AD. Dental biofilms: difficult therapeutic targets. Periodontol 2000 2002;28:12-55.
- Bodet C, Chandad F, Grenier D. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. Micro Infect 2006;8:27-35.
- Beikler T, Ehmke B, Wittstock M, Schmidt H, Karch H, Flemmig TF. Serum antibody reactivity against recombinant PrtC of Porphyromonas gingivalis following periodontal therapy. J Periodont Res 2003;38:276-81.
- Chen LL, Wu YM, Yan J, Sun WL, Sun YZ, Ojcius D. Association between coinfection of Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and Treponema denticola and periodontal tissue destruction in chronic periodontitis. Chin Med J (Engl) 2005;118:915-21.
- Cortelli JR, Cortelli SC, Jordan S, Haraszthy VI, Zambon JJ. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J Clin Periodontol 2005;32:860-6.
- Haffajee AD, Socransky SS, Smith C, Dibart S. Relation of baseline microbial parameters to future periodontal attachment loss. J Clin Periodontol 1991;18:744-50.
- Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000 1999;20:168-238.
- Bedi GS, Williams T. Purification and characterization of a collagen-degrading protease from Porphyromonas gingivalis. J Biol Chem 1994;269:599-606.
- Takahashi N, Kato T, Kuramitsu HK. Isolation and preliminary characterization of the Porphyromonas gingivalis prtC gene expressing collagenase activity. FEMS Micobiol Lett 1991;84:135-8.
- Kato S, Takahashi N, Kuramitsu HK. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J Bacteriol 1992;174:3889-95.
- Norris JM, Love DN. The association of two recombinant proteinases of a feline strain of Porphyromonas gingivalis with periodontal disease in cats. Vet Microbiol 2000;71:69-80.
- Tanahashi T, Kita M, Kodama T, Yamaoka Y, Sawai N, Ohno T, et al. Cytokine expression and production by purified Helicobacter pylori urease in human gastric epithelial cells. Infect Immun 2000;68:664-71.
- Papapanou PN, Sandros J, Lindberg K, Duncan MJ, Niederman R, Nannmark U. Porphyromonas gingivalis may multiply and advance within stratified human junctional epithelium in vitro. J Periodont Res 1994;29:374-5.
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun 1995;63:3878-85.
- Zhou J, Windsor L. Porphyromonas gingivalis affects host collagen degradation by affecting expression, activation, and inhibition of matrix metalloproteinases. J Periodont Res 2006;41:47-54.
- Walter C, Zahlten J, Schmeck B, Schaudinn C, Hippenstiel S, Frisch E, et al. Porphyromonas gingivalis strain-dependent activation of human endothelial cells. Infect Immun 2004;72:5910-8.
- Hack CE, Zeerleder S. The endothelium in sepsis: source of and a target for inflammation. Crit Care Med 2001;29:S21-7.
- Zhou Q, Desta T, Fenton M, Graves DT, Amar S. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis,its lipopolysaccharide, or its FimA protein. Infect Immun 2005;73:935-43.
- Yang QW, Zhu PF, Wang ZG, Jiang JX. Lipopolysaccharide upregulates the expression of Toll-like receptor 4 in human vascular endothelial cells. Chin Med J (Engl) 2002;115:286-9.
- Dong YG, Chen DD, He JG, Guan YY. Effects of 15-deoxy-Δ12,14-prostaglandin J2 on cell proliferation and apoptosis in ECV304 endothelial cells. Acta Pharmacol Sin 2004;25:47-53.
- Jiang JL, Jiang DJ, Tang YH, Li NS, Deng HW, Li YJ. Effect of simvastatin on endothelium-dependent vaso-relaxation and endogenous nitric oxide synthase inhibitor. Acta Pharmacol Sin 2004;25:893-901.
- Armitage GC. Development of a classification system for periodontal disease and conditions. Ann Periodontol 1999;4:1-6.
- Slots J, Ashimoto A, Flynn MJ, Li G, Chen C. Detection of putative pathogens in subgingival specimens by 16S ribosomal DNA amplification with the polymerase chain reaction. Clin Infect Dis 1995;20 Suppl 2:304-7.
- Bodinka AH, Schmidt B, Henkel TF, Flemmig TF, Klaiber B, Karch H. Polymerase chain reaction for the identification of Porphyromonas gingivalis collagenase genes. Oral Microbiol Immunol 1994;9:161-5.
- Slots J, Flynn MJ, Li G. Polymerase chain reaction analysis of the Porphyromonas gingivalis collagenase gene. Clin Infect Dis 1995;20 Suppl 2:S167-8.
- Kuramitsu HK, Yoneda M, Madden T. Proteases and collagenases of Porphyromonas gingivalis. Adv Dent Res 1995;9:37-40.
- Lamont RL, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Molec Biol Rev 1998;62:1244-63.
- Innocenti M, Thoreson AC, Ferrero RL, Stromberg E, Bolin I, Eriksson L, et al. Helicobacter pylori-induced activation of human endothelial cells. Infect Immun 2002;70:4581-90.
- Lee SW, Song KE, Shin DS, Ahn SM, Ha ES, Kim DJ, et al. Alterations in peripheral blood levels of TIMP-1, MMP-2, and MMP-9 in patients with type-2 diabetes. Diabetes Res Clin Pract 2005;69:175-9.
- Chen LL, Yan J. Porphyromonas gingivalis lipopolysaccharide activated bone resorption of osteoclasts by inducing IL-1, TNF, and PGE. Acta Pharmacol Sin 2001;22:614-8.
- Sigusch BW, Guntsch A, Pfitzner A, Glockmann E. Enhanced root planning and systemic metronidazole administration improve clinical and microbiological outcomes in a two-step treatment procedure. J Periodontol 2005;76:991-7.
- Loesche WJ, Grossman NS. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin Microbiol Rev 2001;14:727-52.
- Serino G, Rosling B, Ramberg P, Hellstrom MK, Socransky SS, Lindhe J. The effect of systemic antibiotics in the treatment of patients with recurrent periodontitis. J Clin Periodontol 2001;28:411-8.
- Kasuga Y, Ishihara K, Okuda K. Significance of detection of Porphyromonas gingivalis, Bacteroides forsythus and Treponema denticola in periodontal pockets. Bull Tokyo Dent Coll 2000;41:109-17.
