Dissecting role of regulatory factors in NF-κB pathway with siRNA1
Introduction
The NF-κB family is comprised of a variety of homo- and hetero-dimers formed by p50, p52, RelA (p65), RelB, and c-Rel subunits[1–4]. The best-described form of NF-κB is constituted by the p50 and p65 heterodimer. This heterodimer is sequestered in the cytoplasm bound to a family of inhibitory proteins known as IκB[5,6]. Following cell stimulation, the IκB proteins become phosphorylated by IκB kinase (IKK), a large kinase complex consisting of 2 catalytic subunits, IKKα and IKKβ, and the regulatory subunit, IKKγ/NEMO[7–9]. Phosphorylation of IκB targets this inhibitor for ubiqui-tination and degradation, which results in the release and subsequent translocation of NF-κB to the nucleus to activate transcription of a variety of genes[10] (Figure 1). In this review, we first discuss the current understanding of why RNA-mediated gene silencing by small interfering RNA (siRNA) is important in NF-κB pathway and then focus on the use of siRNA to analyze the role of cellular factors in regulating the NF-κB pathway and its potential use as a targeted therapy to inhibit the NF-κB pathway.

The NF-κB pathway: mechanisms leading to activation
NF-κB can be activated by a variety of stimuli, including inflammatory cytokines, such as TNF-α and IL-1, and growth factors as a result of stress response. Intra-cellular events such as DNA damage by radiation or chemotherapy serve as potent stimulus to activate NF-κB as well. TNF-α and IL-1 are important to the generation of a systemic and local response to infection, injury, and immunological challenges[2]. The signals from the TNF receptor (TNFR) and IL-1 receptor (IL-1R) are transduced through the TNF receptor-associated factor2 (TRAF2) and 6 (TRAF6), respectively[11]. These TRAF are believed to function ‘upstream’ of the cascades of IKK and NF-κB[12,13]. Many members of the mitogen-activated protein kinase kinase kinase (MAP3K) family including MEKK1[14], MEKK2, MEKK3[15], TGF-β-activating kinase1 (TAK1)[16] and NF-κB-inducing kinase (NIK)[17] also activate IKK when overexpressed. However, MEKK3 is an essential signal transducer in both TNFR- and IL-1R-induced NF-κB activation[11,18-22].
Initial studies on the structure of TRAF showed that the C-terminal domain for TRAF was responsible for protein-protein interactions and the N-terminal region of TRAF, including a RING finger and a variable number of Zn finger domains, was necessary for TRAF mediated activation of downstream signaling pathways[23,24]. A RING finger with ubiquitin ligase activity[25] is critical for NF-κB activation by TRAF[26–28]. TRAF2 and TRAF6 were shown to be able to function as ubiquitin ligases that autoubiquitinate resulting in a lysine-63 (K63)-linked polyubiquitination[29–31]. Ubiqui-tinated TRAF can bind to the TAK1, and its adapter proteins TAB1 and TAB2[18,32,33]. TAB1 binds to TAK1 and is involved in regulating its activity, while TAB2 binds preferentially to K63-linked polyubiquitin chains[12], resulting in the activation of TAK1[34]. Activated TAK1 can phosphorylate IKK directly or act on the NF-κB-inducing kinase (NIK), which in turn activates IKK[16,35]. .Thus, the TAK1 complex is an important link between TRAF and the NF-κB pathway (Figure 1). TRAF7 also plays an important role in regulating activation of NF-κB and it can act like TRAF6 in relaying signals and activating the NF-κB pathway[18].
IKK activation by the TNF-α and IL-1 is a rapid, but transient process, implying a negative feedback regulation of IKK following its activation. This negative regulation of IKK is controlled, at least in part, by deubiquitination, as shown in recent studies on the tumor suppressor cylindro-matosis protein CYLD[36,37]. Loss of CYLD has been linked to a predisposition to cylindromas, a syndrome characterized by benign tumors of the skin appendages. Interestingly, CYLD contains cysteine and histidine boxes found in the ubiquitin-specific protease (UBP) family of deubiquitination enzymes[38]. Moreover, a portion of the histidine box of CYLD is deleted in some cylindromatosis patients, suggesting a link between the deubiquitination activity of CYLD and its tumor suppressor function. Three independent studies have shown that CYLD binds to NEMO and facilitates the disassembly of K63-linked polyubiquitin chains on TRAF2 and TRAF6[36,37]. Thus, a critical function of CYLD is to down-regulate NF-κB activation through its deubiquitinating activity[31].
RNAi-mediated gene silencing
RNAi is associated with a number of practical and theoretic advantages over pre-existing methods of suppressing gene expression (Table 1)[39–44] and thus provides a useful mean to dissect the role of various factors that regulate the NF-κB pathway. RNAi also has the potential to be developed as a therapeutic modality to knock-down gene products that are important in activating the NF-κB pathway[45]. Several lines of evidence support a role for RNAi in a cell-based defense mechanism that protects the genome against mobile genetic elements such as viruses and transposons[46,47]. There are 2 classes of small RNA that can silence gene expression. One class is processed from double-stranded (dsRNA) precursor molecules into small interfering RNA (siRNA) by the RNAase III-like nuclease called Dicer; these siRNA act as guides for the siRNA-induced silencing complex (siRISC) to target and cleave complementary mRNA[48]. Dicer processes another class of small RNA from pre-microRNA into microRNA. These micro RNA act as guides for a multiprotein complex (miRISC) which identifies mRNA and silences gene expression either via destruction of the mRNA or by blocking its translation[47,49,50]. Dicer was first isolated from extracts of Drosophila, but was later shown to exist in a large variety of species ranging from fungi to man[51]. Two Dicers, Dcr-1 and Dcr-2 were found in Drosophila. Dcr-1 processes pre-miRNA[52,53], while Dcr-2 processes dsRNA. In contrast to their processing specificities, both Dcr-1, Dcr-2 and its associated factor R2D2 are required for assembly of siRNA into siRISC (Figure 2)[47,54]. Synthetic 21-23 nucleotide double stranded siRNA were synthesized to resemble Dicer cleavage products and could be directly incorporated in the mammalian RISC to target mRNA for degradation[55]. Another approach relies on stable expression of short hairpin RNA from a plasmid vector down stream from a pol III or U6 promoter to result in a reproducible reduction of target gene expression in mammalian cells[45]. Various strategies including retroviral, adenoviral and lentiviral vectors have been developed that allow the introduction of siRNA encoding vectors at high efficiency in primary cells. With these technologies, it is now possible to obtain effective gene silencing in transgenic embryos and adult mice[48]. There have been several reports of successful use of in vivo siRNA in different animal models of human diseases; for example, microinjection of siRNA directed against zebrafish dystrophin gene into zebrafish embryos demonstrates the efficacy of siRNA-based gene silencing in this model and illustrates the potential of this approach to determine the roles of multiple protein products expressed by a single gene during the early stages of development[56]. Delivery of siRNA directed against either caspase 8 or hepatitis B virus (HBV) by mouse tail vein have been effective in suppressing specific gene expression[57,58]. In addition, Verma et al demonstrated that siRNA directed against β-catenin reduced tumor growth in nude mice when administered by either intravenous or intraperitoneal injections, which suggests that siRNA could have therapeutic potential for inhibiting the expression of genes that enhance the growth of tumors[59]. RNAi also holds great promise for the treatment of CNS diseases in which neurodegeneration is linked to overproduction of endogenous protein or to synthesis of aberrant proteins coded by dominant mutant alleles[60].
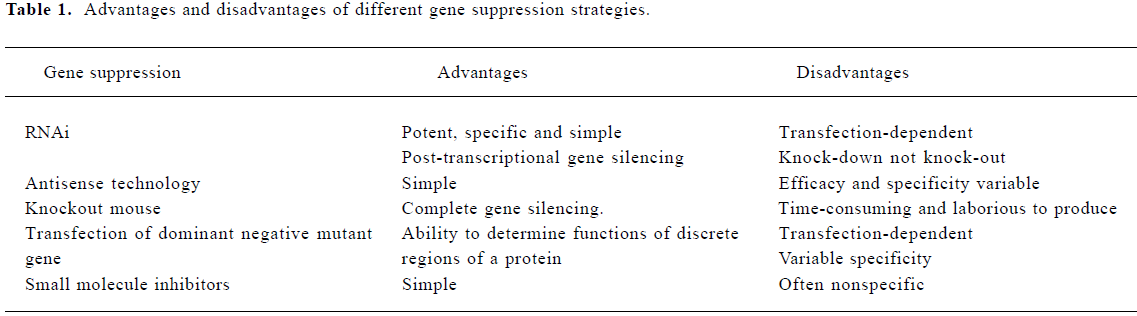
Full table

More recently, many researchers have used plasmid and viral vectors for transcription of short-hairpin RNA (shRNA) that efficiently deliver siRNA into both dividing and non-dividing cells, stem cells, zygotes, and their differentiated progeny. Gene expression was more stably inhibited with these expression systems than with the transient knockdown recorded with chemically synthesised siRNA. A number of groups have used shRNA instead of siRNA to obtain relatively long-lived gene silencing in vivo[61]. The libraries of retroviruses expressing shRNA designed to silence large fractions of all expressed human genes have been produced. These shRNA libraries have the potential to provide mammalian biologists for the first time with a genetic screening tool similar to that which has been used in more primitive organisms.
RNAi is an important tool for analyzing the NF-κB pathway
Signal transduction pathways, such as the NF-κB pathway, are modular composites of functionally interdependent sets of proteins that act in a coordinated fashion to transform environmental information into a phenotypic response. Several mechanisms that cause constitutive NF-κB activity can be found in different epithelial tumors, tumor cell lines and lymphoid malignancies[62]. Many different inhibitors affecting the NF-κB activation pathway that have beneficial effects on tumor development or that increase the response to radiation and chemotherapy have been described. However, most of these inhibitors are not specific and inhibit many other pathways as well[63]. Using RNAi to stably knock out specific gene expression and function is a highly effective and novel method that is rapidly gaining ground because of an explosion of new and improved techniques. The exquisite sequence specificity of RNAi provides a promising approach to address the complex interactions of viral and cellular regulatory proteins involved in NF-κB pathway[64]. As potent small-molecule inhibitors to any gene expression, siRNA can be used to specifically analyze the role of single gene products in NF-κB pathway to emphasize the selectivity of RNAi-based therapy. Direct evidence, using both in vitro and in vivo models, indicates that RNAi is a critical tool to inhibit the NF-κB pathway at multiple levels and study the transmission of signals in both physiologic and pathologic states[36,37,65–68].
TAK1 is an important upstream mediator of the NF-κB pathway[16,34,46]. siRNA directed against TAK1 decreased the amount of both IL-1 and TNFα-induced phospho-IκBα expression and prevented IκBα degradation. The loss of endogenous TAK1 by siRNA resulted in impaired DNA-binding of NF-κB. These results provide the first genetic evidence that supports a role of TAK1 as a critical upstream kinase for IKKα or IKKβ in IL-1 and TNFα-induced activation of the NF-κB pathway[69]. Takaesu et al also reported that endogenous IKKα and IKKβ co-immunoprecipitated with TAK1 upon TNFα stimulation. siRNA directed against IKKα and IKKβ reduced IL-1 and TNFα-induced activation of the NF-κB pathway. Simultaneous transfection of both IKKα and IKKβ siRNA resulted in further decreases in NF-κB activation as compared to transfection of each of these individual siRNA. These findings suggest that in addition to IKKα and IKKβ, TAK1 is important for NF-κB activation challenging previous result that only IKKβ was involved in NF-κB activation[28,70–72].
Bcl-10, a cellular homolog of the equine herpesvirus-2 E10 gene[73], was found over-expressed in some lymphomas of the mucosa-associated lymphoid tissue (MALT). Bcl-10 has been shown to physically associate with MALT1, which is a member of the paracaspase family and also involved in MALT lymphoma. Bcl-10 and MALT1 are essential for the activation of IKK and NF-κB in response to T cell receptor stimulation[74]. Sun et al presented evidence that TRAF2 and TRAF6 mediated IKK activation by Bcl-10 and T cell receptor stimulation. TRAF6 siRNA reduced the activation of IKK by 50% and the same percentage of reduction in IKK activation was observed by TRAF2 siRNA. However, the combination of TRAF2 and TRAF6 siRNA reduced IKK activation by approximately 80%. Thus in T cells, both TRAF2 and TRAF6 are involved in upstream regulation of the NF-κB pathway in response to T cell receptor stimulation[64]. In addition, TAK1 siRNA transfection also dramatically reduced IKK activation by T cell receptor stimulation in T cells. Moreover, MALT1 and Bcl-10 have been shown to mediate IKK activation by facilitating the K63 polyubiquitination of NEMO. siRNA that reduced the expression of paracaspase and Ubc13 abrogated the effects of Bcl-10, which indicates that Bcl-10 promotes activation of IKK and NF-κB through paracaspase- and Ubc13-dependent ubiquitination of NEMO[75].
The tumor suppressor cylindromatosis protein CYLD belongs to a subfamily of enzymes with deubiquitinase activity[76,77]. A collection of shRNA that suppress 50 human de-ubiquitinating enzymes were used to identify deubiqui-tination enzymes and study the mechanism for human cylindromatosis[36] in the NF-κB pathway[45]. The studies from this and other groups show that CYLD binds to NEMO, and appears to regulate its activity through deubiquitination of TRAF2[38,78]. They also demonstrated that inhibition of CYLD by siRNA enhanced NF-κB activation and prevented apoptosis, suggesting a mechanism through which loss of CYLD contributes to oncogenesis[36]. In independent studies, Trompouki et al[37] also used the siRNA method and demonstrated that CYLD interacted with NEMO and negatively regulated NF-κB signaling by deubiquitination of TRAF2. They have now started to investigate the use of CYLD inhibitors in clinical trials.
Potential therapeutic uses of NF-κB inhibition by siRNA
The NF-κB signaling pathway is important in the generation of the monocyte-derived dendritic cells and regulates their functional maturation and activation[79–81]. Dendritic cells play a prominent role in infectious diseases, immune disorders, and in cancer immunology[82]. In mammalian cells, NF-κB/Rel proteins are involved in regulating survival, differentiation, and activation of the dendritic cells[80,81,83]. Targeted mutations in mice demonstrate that deficits in RelB, cRel, p50, or p52 lead to various immune impairments that directly implicate dendritic cells. Transfection of dendritic cells with p50 siRNA was tested by Diego[88] and his colleagues as a way of performing loss-of-function analysis in vitro and the results showed strong and specific down-regulation of both p50 mRNA and protein levels. Such interference impaired p50 nuclear localization and DNA-binding in response to CD40 Ligand (CD40L) and IL-1 activation. The cytosolic fraction also showed reduced p50 activity after p50 siRNA transfection[88].
IL-12 is a cytokine pivotal for the development of cellular immunity and production of high levels of IFN-γ by T cells. A biologically active form of IL-12 (IL-12α and IL-12β heterodimer) is produced from the transcription of separate genes which are regulated independently[89]. Prior results have shown that CD40L alone or in combination with IL-1 induces high levels of IL-12β transcription[90]. However, a significantly reduced IL-12β mRNA level and reduction of the secretion of IL-12αβ heterodimer was observed after p50 siRNA, which suggests that p50 siRNA down-regulated the production of IL-12 in response to CD40L and IL-1. These results are consistent with studies of the promoter of the IL-12β gene, which is NF-κB inducible and contains sites for the binding of p50 in B cells[91].
It has been reported that p65 can stimulate HIV-1 transcriptional elongation by binding to the HIV-1 long terminal repeat (LTR)[92,93]. The use of siRNA directed against p65 resulted in reduced HIV-1 replication, which correlated with the decrease in HIV-1 virons in supernatants from MAGI cells[66]. CD4-positive human T-lymphocyte cell lines including MAGI have been used to study different aspects of the HIV-1 life cycle. These cells, which stably express CD4 receptors on the cell surface, can be infected by HIV-1. Since they contain a HIV-1 LTR fused to the β-galactosidase gene, infectious virus can transactivate the LTR-β-galactosidase reporter and increase β-galactosidase activity. Thus, staining of MAGI cells to determine β-galactosidase activity makes these cells an excellent indicator to determine the number of HIV-1 infectious particles[94]. It has been observed that more than 90% of the infected cells transfected with control siRNA demonstrated marked β-galactosidase positivity. In contrast, only a few cells had β-galactosidase activity when the MAGI cells were transfected with p65 siRNA, which indicate that inhibition of HIV-1 replication by p65 siRNA resulted in very low levels of HIV-1 infectious particles[66]. This finding highlights the importance that NF-κB plays in the life cycle of HIV-1.
Tumors that have constitutive NF-κB activity show increased resistance to chemotherapy. Inhibition of NF-κB does not only lead to enhanced apoptosis but also to increased sensitivity to radiation or chemotherapy in several tumor cells such as fibrosarcoma and colorectal cancer cell lines, as well as xenograft models or pancreatic carcinoma cells[63]. CPT-11 is a topoisomerase I inhibitor which has efficacy in the treatment of certain neoplasms including colorectal cancer. In spite of the initial response to therapy[81], most tumors from patients treated with CPT-11 become resistant and exhibit tumor progression[95]. However, inducible chemotherapy resistance to CPT-11 has been shown to be reversed by inhibiting NF-κB[83,96]. More recent studies from Guo et al[65] demonstrate that NF-κB activation induced by CPT-11 in the relatively resistant HCT116 cell line is effectively inhibited by p65 siRNA both in vitro and in vivo. Transfection of p65 siRNA into HCT116 cells dramatically reduced the expression of p65. In addition, they found that loss of p65 did not impact cell viability on its own, but p65 siRNA in conjunction with CPT-11 increased tumor cell sensitivity to the cytotoxic effects of CPT-11. P65 siRNA increased apoptosis and reduced NF-κB-binding activity. The effect on apoptosis could be partly explained by down-regulation of the NF-κB target genes c-IAP1 and c-IAP2[65]. These results are consistent with the role that NF-κB plays in the inhibition of CPT-11 mediated cell killing[2,97]. Importantly, transient exposure of HCT116 cells to p65 siRNA in cell culture altered the ability of these cells to proliferate following injection into nude mice in the presence of CPT-11 treatment. Systemic therapy with intravenous injection of p65 siRNA did not limit tumor growth. However, when combined with CPT-11, intravenous injection of p65 siRNA significantly delayed tumor growth with dramatic reductions in tumor volume[65]. These studies demonstrate that delivery of siRNA to tumor cells in vivo is feasible and that inhibition of NF-κB-mediated transcription by p65 siRNA holds therapeutic promise in cancer[98].
Questions to the safety and efficacy of using RNAi as a therapeutic strategy
Interest in RNAi initially was restricted to basic researchers to study gene function. The subsequent finding that in vivo delivery of siRNA to induce RNAi in mammalian cells has generated excitement regarding its potential therapeutic applications. Various approaches have been shown to improve cell and tissue delivery of siRNA and shRNA[61,99].
A major obstacle to the development of siRNAi as a therapeutic tool is its delivery to the desired cell type in the correct tissue or organ. Hydrodynamic delivery of siRNA that involves the intravascular injection of large fluid volumes in order to locally increase intravascular pressure[100,101] might be adapted for local administration of siRNA by arterial or venous catheters in organs such as liver, kidney, heart or lungs, but cannot be utilized for systemic treatment. Intravenous injection of siRNA in large volumes of saline solution, works by creating a back-flow in the venous system that forces the siRNA solution into several organs with lesser efficiency[102,103]. Using RNAi to silence genes is also limited by the stability of siRNA molecules in vivo and the efficiency with which they are taken up by target cells and tissues[104]. An additional obstacle in exploring siRNA as a therapeutic tool is toxicity. siRNA have the potential to induce a concentration- and cell-type-dependent cell death[105]. In mammalian cells, the utility of RNAi has been limited by the innate immune response triggered by dsRNA. Long dsRNA induce an interferon response usually resulting in a generalized inhibition of gene expression. However, this response can usually be avoided in mammalian cell cultures by using synthetic siRNA with a length of 21 nt[105].
Inhibition of viral replication by RNAi has been demonstrated in vitro for a variety of viruses, including RNA viruses such as HIV, respiratory syncytial virus, influenza virus, poliovirus, West Nile virus, dengue virus, and foot and mouth disease virus. However, some viruses are resistant to RNAi; for example, although siRNA can inhibit the production of progeny virus, the genomic RNA of respiratory syncytial virus, hepatitis delta virus, and rotavirus are resistant to RNAi, either because of tight shielding by proteins or to sequestration in compartments inaccessible to siRNA[106–108]. Moreover, some viruses such as influenza and vaccinia produce proteins that actively suppress silencing by RNAi[109]. Adenovirus was recently shown to block the processing of shRNA in mammalian cells by expressing a viral noncoding RNA at such high levels that it binds most of the available RNAi processing machinery[110].
Summary
The discovery of RNAi has already provided a powerful tool for basic science researchers to study gene function. More recently the use of RNAi for genetic-based therapies has been widely studied, especially in viral infections, cancers, and inherited genetic disorders. Combined with genomics data, RNAi-directed gene-silencing could allow functional determination of any gene expressed in a cell or pathway. Thus, the therapeutic potential for RNAi is enormous, but the ability to efficiently and stably produce and deliver sufficient amounts of siRNA to the target tissues require refinement before this new technology can be tried clinically[111].
Acknowledgments
We thank Richard GAYNOR and Udit VERMA for their helpful comments during the preparation of the manuscript. We also thank Janice BOX for the preparation of the manuscript and Alejandra HERRERA for graphical support.
References
- Perkins ND. The Rel/NF-kappa B family: friend and foe. Trends Biochem Sci 2000;25:434-40.
- Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest 2001;107:241-6.
- Baeuerle PA, Baltimore D. κB: Ten years after. Cell 1996;87:13-20.
- Ghosh S, May MJ, Kopp EB. κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998;16:225-60.
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell 2002;109:S81-S96.
- Verma IM, Stevenson JK, Schwartz EM, Van Antwerp D, Miyamoto S. κB/IκB family: intimate tales of association and dissociation. Genes Dev 1995;9:2723-35.
- DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. κB kinase that activates the transcription factor NF-κB. Nature 1997;388:548-54.
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, et al. κB kinases essential for NF-κB activation. Science 1997;278:860-6.
- Li Y, Kang J, Friedman J, Tarassishin L, Ye J, Kovalenko A, et al. κB activity and as a target of an adenovirus inhibitor of tumor necrosis factor α-induced apoptosis. Proc Natl Acad Sci USA 1999;96:1042-7.
- Stancovski I, Baltimore D. NF-kappaB activation: the I kappaB kinase revealed? Cell 1997;91:299-303.
- Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev 1999;13:1297-308.
- Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 2004;16:339-40.
- Wajant H, Scheurich P. Analogies between Drosophila and mammalian TRAF pathways. Prog Mol Subcell Biol 2004;34:47-72.
- Lee FS, Peters RT, Dang LC, Maniatis T. MEKK1 activates both IkappaB kinase alpha and IkappaB kinase beta. Proc Natl Acad Sci USA 1998;95:9319-24.
- Zhao Q, Lee FS. Mitogen-activated protein kinase/ERK kinase kinases 2 and 3 activate nuclear factor-kappaB through IkappaB kinase-alpha and IkappaB kinase-beta. J Biol Chem 1999;274:8355-8.
- Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 1999;398:252-6.
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. κB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science 1997;278:866-9.
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 2004;6:97-105.
- Malinin NL, Boldin MP, Kovalenko AV, Wallach D. κB induction by TNF, CD95 and IL-1. Nature 1997;385:540-8.
- Song HY, Regnier CH, Kirschning CJ, Goeddel DV, Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-kappaB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci USA 1997;94:9792-6.
- Yang J, Lin Y, Guo Z, Cheng J, Huang J, Deng L, et al. The essential role of MEKK3 in TNF-induced NF-kappaB activation. Nat Immunol 2001;2:620-4.
- Huang Q, Yang J, Lin Y, Walker C, Cheng J, Liu ZG, et al. Differential regulation of interleukin 1 receptor and Toll-like receptor signaling by MEKK3. Nat Immunol 2004;5:98-103.
- Cheng G, Cleary AM, Ye ZS, Hong DI, Lederman S, Baltimore D. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science 1995;267:1494-8.
- Rothe M, Sarma V, Dixit VM, Goeddel DV. κB by TNF receptor 2 and CD40. Science 1995;269:1424-7.
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem 2001;70:503-33.
- Takeuchi M, Rothe M, Goeddel DV. Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J Biol Chem 1996;271:19935-42.
- Dadgostar H, Cheng G. An intact zinc ring finger is required for tumor necrosis factor receptor-associated factor-mediated nuclear factor-kappaB activation but is dispensable for c-Jun N-terminal kinase signaling. J Biol Chem 1998;273:24775-80.
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, et al. A subunit of IκB kinase. Science 1999;284:316-20.
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000;103:351-61.
- Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal 2001;13:389-400.
- Wilkinson KD. Signal transduction: aspirin, ubiquitin and cancer. Nature 2003;424:738-9.
- Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, et al. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science 1996;272:1179-82.
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 2001;412:346-51.
- Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell 2000;5:649-58.
- Shirakabe K, Yamaguchi K, Shibuya H, Irie K, Matsuda S, Moriguchi T, et al. TAK1 mediates the ceramide signaling to stress-activated protein kinase/c-Jun N-terminal kinase. J Biol Chem 1997;272:8141-4.
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003;424:797-801.
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 2003;424:793-6.
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet 2000;25:160-5.
- Dave RS, Pomerantz RJ. RNA interference: on the road to an alternate therapeutic strategy. Rev Med Virol 2003;13:373-85.
- Schmidt CW. Therapeutic interference: small RNA molecules act as blockers of disease metabolism. Modern Drug Disc 2003: 37–42.
- Arenz C, Schepers U. RNA interference: from an ancient mechanism to a state of the art therapeutic application? Naturwis-senschaften 2003;90:345-59.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806-11.
- Hohmann HP, Brockhaus M, Baeuerle PA, Remy R, Kolbeck R, van Loon AP. Expression of the types A and B tumor necrosis factor (TNF) receptors is independently regulated, and both receptors mediate activation of the transcription factor NF-kappa B. TNF alpha is not needed for induction of a biological effect via TNF receptors. J Biol Chem 1990;265:22409-17.
- Boutla A, Delidakis C, Livadaras I, Tsagris M, Tabler M. Short 5'-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr Biol 2001;11:1776-80.
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550-3.
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature 2001;411:834-42.
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A. Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 2004;117:83-94.
- Medema RH. Optimizing RNA interference for application in mammalian cells. Biochem J 2004;380:593-603.
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004;117:69-81.
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol 2004;16:223-9.
- Hannon GJ. RNA interference. Nature 2002;418:244-51.
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002;297:2056-60.
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev 2003;17:438-42.
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003;301:1921-5.
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev 2005;19:517-29.
- Dodd A, Chambers SP, Love DR. Short interfering RNA-mediated gene targeting in the zebrafish. FEBS Lett 2004;561:89-93.
- Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci USA 2003;100:7797-802.
- Klein C, Bock CT, Wedemeyer H, Wustefeld T, Locarnini S, Dienes HP, et al. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology 2003;125:9-18.
- Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res 2003;9:1291-300.
- Forte A, Cipollaro M, Cascino A, Galderisi U. Small interfering RNAs and antisense oligonucleotides for treatment of neurological diseases. Curr Drug Targets 2005;6:21-9.
- Zhang Y, Boado RJ, Pardridge WM. In vivo knockdown of gene expression in brain cancer with intravenous RNAi in adult rats. J Gene Med 2003;5:1039-45.
- Gilmore T, Gapuzan ME, Kalaitzidis D, Starczynowski D. Rel/NF-kappa B/I kappa B signal transduction in the generation and treatment of human cancer. Cancer Lett 2002;181:1-9.
- Greten FR, Karin M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett 2004;206:193-9.
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell 2004;14:289-301.
- Guo J, Verma UN, Gaynor RB, Frenkel EP, Becerra CR. Enhanced chemosensitivity to irinotecan by RNA interference-mediated down-regulation of the nuclear factor-kappaB p65 subunit. Clin Cancer Res 2004;10:3333-41.
- Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J Virol 2002;76:12963-73.
- Brisibe EA, Okada N, Mizukami H, Okuyama H, Fujii YR. RNA interference: potentials for the prevention of HIV infections and the challenges ahead. Trends Biotechnol 2003;7:306-11.
- Lee NS, Rossi JJ. Control of HIV-1 replication by RNA interference. Virus Res 2004;102:53-8.
- Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB. TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J Mol Biol 2003;326:105-15.
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, et al. Science 1999;284:313-6.
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. κB kinase 2 gene. Science 1999;284:321-5.
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, et al. kB kinase (IKK) is essential for nuclear factor kappa B activation and prevention of apoptosis. J Exp Med 1999;189:1839-45.
- Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 1999;96:35-45.
- Lucas PC, McAllister-Lucas LM, Nunez G. NF-kappaB signaling in lymphocytes: a new cast of characters. J Cell Sci 2004;117:31-9.
- Zhou H, Wertz I, O’Rourke K, Ultsch M, Seshagiri S, Eby M, et al. Bcl10 activates the NF-kappaB pathway through ubiquitina-tion of NEMO. Nature 2004;427:167-71.
- Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J 1997;11:1245-56.
- D’Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol 1998;33:337-52.
- Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science 2002;296:1634-5.
- Yoshimura S, Bondeson J, Brennan FM, Foxwell BM, Feldmann M. Role of NFkappaB in antigen presentation and development of regulatory T cells elucidated by treatment of dendritic cells with the proteasome inhibitor PSI. Eur J Immunol 2001;31:1883-93.
- Yoshimura S, Bondeson J, Foxwell BM, Brennan FM, Feldmann M. Effective antigen presentation by dendritic cells is NF-kappaB dependent: coordinate regulation of MHC, co-stimulatory molecules and cytokines. Int Immunol 2001;13:675-83.
- O’Sullivan BJ, Thomas R. CD40 ligation conditions dendritic cell antigen-presenting function through sustained activation of NF-kappaB. J Immunol 2002;168:5491-8.
- Silverman N, Maniatis T. κB signaling pathways in mammalian and insect innate immunity. Genes Dev 2001;15:2321-42.
- Neumann M, Fries H, Scheicher C, Keikavoussi P, Kolb-Maurer A, Brocker E, et al. Differential expression of Rel/NF-kappaB and octamer factors is a hallmark of the generation and maturation of dendritic cells. Blood 2000;95:277-85.
- Burkly L, Hession C, Ogata L, Reilly C, Marconi LA, Olson D, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 1995;373:531-6.
- Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev 1995;9:1965-77.
- Sha WC, Liou HC, Tuomanen EI, Baltimore D. κB leads to multifocal defects in immune responses. Cell 1995;80:321-30.
- Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, et al. κB/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med 1998;187:147-59.
- Laderach D, Compagno D, Danos O, Vainchenker W, Galy A. RNA interference shows critical requirement for NF-kappa B p50 in the production of IL-12 by human dendritic cells. J Immunol 2003;171:1750-7.
- Trinchieri G. Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv Immunol 1998;70:83-243.
- Wesa AK, Galy A. IL-1 beta induces dendritic cells to produce IL-12. Int Immunol 2001;13:1053-61.
- Gri G, Savio D, Trinchieri G, Ma X. Synergistic regulation of the human interleukin-12 p40 promoter by NFkappaB and Ets transcription factors in Epstein-Barr virus-transformed B cells and macrophages. J Biol Chem 1998;273:6431-8.
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. κB binds P-TEFβ to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 2001;8:327-37.
- West MJ, Lowe AD, Karn J. Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol 2001;75:8524-37.
- Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galac tosidase gene. J Virol 1992;66:2232-9.
- Hannon GJ, Conklin DS. RNA interference by short hairpin RNAs expressed in vertebrate cells. Methods Mol Biol 2004;257:255-66.
- Hilliard B, Samoilova EB, Liu TS, Rostami A, Chen Y. Experimental autoimmune encephalomyelitis in NF-kappa B-deficient mice: roles of NF-kappa B in the activation and differentiation of autoreactive T cells. J Immunol 1999;163:2937-43.
- Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS Jr. κB anti-apoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998;281:1680-3.
- Veiby OP, Read MA. Chemoresistance: impact of nuclear factor (NF)-kappaB inhibition by small interfering RNA. Commentary re J. Guo et al. Enhanced chemosensitivity to irinotecan by RNA interference-mediated down-regulation of the NF-kappaB p65 subunit. Clin Cancer Res 2004;10:3333-41.
- Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett 2004;558:63-8.
- Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003;9:347-51.
- Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep 2003;4:602-8.
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature 2002;418:38-9.
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet 2002;32:107-8.
- Dillon CP, Sandy P, Nencioni A, Kissler S, Rubinson DA, Van Parijs L. RNAi as an experimental and therapeutic tool to study and regulate physiological and disease processes. Annu Rev Physiol 2005;67:147-73.
- Achenbach TV, Brunner B, Heermeier K. Oligonucleotide-based knockdown technologies: antisense versus RNA interference. Chembiochemistry 2003;4:928-35.
- Silvestri LS, Taraporewala ZF, Patton JT. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol 2004;78:7763-74.
- Chang J, Provost P, Taylor JM. Resistance of human hepatitis delta virus RNAs to dicer activity. J Virol 2003;77:11910-17.
- Bitko V, Barik S. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol 2001;1:34.
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 2004;101:1350-5.
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 2004;78:12868-76.
- Shankar P, Manjunath N, Lieberman J. The prospect of silencing disease using RNA interference. JAMA 2005;293:1367-73.
