Antisense oligonucleotides-induced local blockade of T-bet expression leads to airway inflammation in rats1
Introduction
Asthma is a chronic pulmonary disease associated with reversible airflow obstruction and chronic inflammation of the airway[1,2]. In asthma, there is a polarization of T-lymphocyte responses and enhanced secretion of cytokines involved in the regulation of immunoglobulin (Ig)-E, mast cells, basophils, and eosinophils, ultimately leading to airway inflammation[3]. The proinflammatory cytokines produced by T cells contribute to the initiation and perpetuation of allergic asthma[4].
In recent years, studies have found that some transcription factors play key roles in the tightly regulated mechanism governing T-helper cell type 1 (Th1), Th2 cell differentiation, and the ongoing maintenance of the corresponding immune response. GATA binding protein 3 (GATA-3) and T-box expressed in T cells (T-bet) are 2 T-cell-specific transcription factors. GATA-3 is a pleiotropic transcription factor of the C4 zinc finger family that binds to a 5'-WGATAR-3' consensus DNA sequence which is found to be selectively expressed in Th2, but not in Th1 cells, and plays an important role in cytokine gene expression in T cells[5,6]. Ectopic expression of GATA-3 in developing T cells leads to the up-regulation of interleukin-4 (IL-4) and IL-5, and the down-regulation of interferon-γ (IFN-γ) in allergic asthma. In a recently published study, Finotto et al found that the local delivery of GATA-3 AS-ODN might be a novel approach by the down-regulation of IL-4, and the up-regulation of IFN-γ for the treatment of airway hyperresponsiveness in asthma[7] which has the potential advantage of suppressing the expression of various proinflammatory Th2 cytokines simultaneously rather than suppressing the activity of a single cytokine.
The other transcription factor, T-bet, a recently discovered transcription factor, is selectively expressed in Th1 cells and plays a critical role in Th1 differentiation[8]. The T-box transcription factors have a consensus binding site, and inspection of the IFN-γ gene reveals 3 potential T-box binding sites, 2 of which are located within the proximal promoter region and the other is located in the third intron[8]. IFN-γ is a pleiotropic cytokine that regulates both differentiation and the stabilization of Th1 cells[9–11] through binding to a receptor composed of the IFN-γR1 and IFN-γR2 subunits. T-bet can induce the expression of IFN-γ. In our recently published study[12], we found that T-bet was down-regulated in asthmatic patients compared with normal subjects. Furthermore, Benjamin et al found that T-bet polymorphisms were associated with airway hyperresponsiveness[13]. Szabo and Glimcher developed T-bet-deficient mice in which CD4+ T lymphocytes produced less IFN-γ and more IL-4 and IL-5 than wild-type mice. Importantly and interestingly, in the absence of any sensitization or allergen challenge, T-bet-deficient mice have airway hyperresponsiveness to metha-choline, and recruit eosinophils and lymphocytes to the airway[14].
Based on this data, it was of particular interest to analyze the expression and functional role of T-bet and GATA-3 genes in asthma. Thus, we hypothesize that antisense-induced local blockade of T-bet expression in the lungs without any sensitization or allergen challenge of ovalbumin (OVA) can lead to airway inflammation.
Materials and methods
Reagents The following reagents were used in the present study: aluminum hydroxide, crystalline OVA (Sigma, St Louis, Missouri, USA), polyclonal rabbit antibody anti-GATA-3, monoclonal mouse antibody anti-T-bet, and polyclonal rabbit antibody anti-beta-actin (Santa Cruz Biotechnology Inc, Santa Cruz, CA, USA), biotinylated anti-goat IgG (Boster, Wuhan, Hubei, China.), IL-4, IL-5, and IFN-γ ELISA kits (Jingmei Biotech, Shenzhen, Guangdong, China), in situ hybridization detection kit for biotin labeled probes of T-bet and GATA-3 (Sigma, St Louis, Missouri, USA), and T-bet antisense phosphorothioate oligonucleotides (Bochai Biotech, Shanghai, China).
Oligonucleotides were synthesized with a phosphorothioate backbone to improve resistance to endonucleases. The AS-ODN consisted of 20-mer analogues to the 5' end of the rat T-bet sequence which spans the translation initiation site. This sequence had no C plus G (CpG) dinucleotide[15] or a quadruple G sequence element that are known to cause unspecific effects. Furthermore, several control (nonsense) oligonucleotides were prepared, and the nonsense oligonucleotides contained the same nucleotide composition as the AS-ODN. The sequences of the biotin labeled probe of T-bet and GATA-3 for in situ hybridization detection, T-bet antisense DNA, and T-bet nonsense DNA are listed in Table 1.
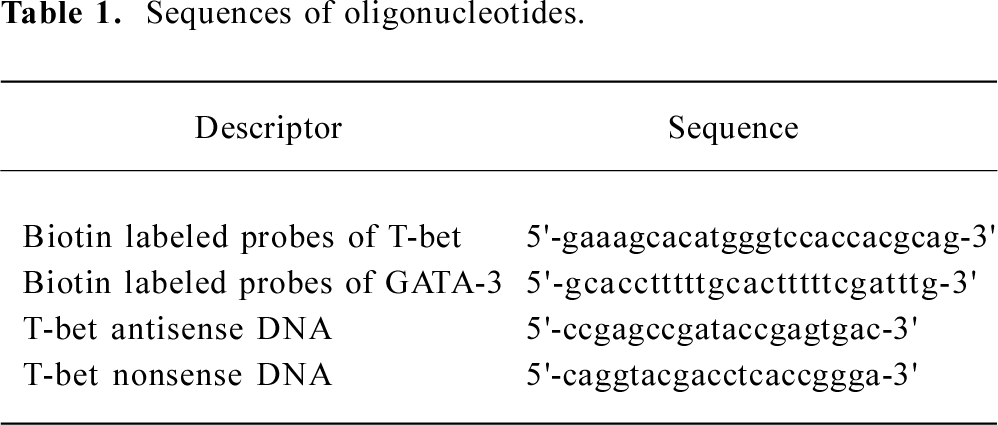
Full table
Animals, allergen sensitization, and study protocol The present study was approved by the Animal Ethics Committees of West China Hospital at Sichuan University. Twenty-four specific pathogen-free, female Wistar rats (8–10 weeks of age) were obtained from the Experimental Animal Center of Sichuan University. The rats kept in standard animal research facilities were given food and water and had filtered air. The rats weighed between 150 and 180 g at the time of testing.
The 24 rats were then randomly divided into 4 groups: saline group, OVA-sensitized group, antisense group, and nonsense group. The 6 rats in the OVA-sensitized group were sensitized by intraperitoneal injection on d 0 and d 7 with 1 mg OVA and 200 µg Al(OH)3 in 0.5 mL saline. From d 14 to d 21, the rats underwent aerosol challenge with OVA (1%, w/v) lasting 30 min per 2 days. The animals in the saline group underwent saline sensitization and aerosol challenge with saline. The 6 rats in the antisense group underwent aerosol delivery of the AS-ODN with 3 mg (0.1%, w/v), and aerosol delivery of nonsense oligonucleotides with 3 mg (0.1%, w/v) was administered intratracheally in the nonsense group from d 0 to d 21 per 2 days.
Bronchoalveolar lavage and IL-4, IL-5, and IFN-γ enzyme-linked immunosorbent assay The left lungs of the rats were used for bronchoalveolar lavage (BAL) analysis. The trachea was cannulated with a 20-gauge catheter that was secured with a ligature, and lavage of the left lung was performed using a 3-mL aliquot of saline (0.9% NaCl at 4 ºC). A total of 3 lavages were performed after tying off the right lung at the mainstem bronchus. In all the rats, the recovery of the lung lavage fluid was 80% or greater. The recovered fluid was centrifuged at 1 200 r/min for 5 min to deposit the cells and the supernatants of BAL fluid (BALF) were immediately placed on melting ice. Four hundred cells of a Wright–Giemsa-stained cytocentrifuge slide preparation were counted for determination of the differential cell recovery. To determine cytokine concentrations in BALF, supernatants were analyzed by specific ELISA for concentration of IL-4, IL-5, and IFN-γ.
Lung histology and in situ hybridization detection for T-bet and GATA-3 mRNA After bronchoalveolar lavage, the right lung was fixed in 10% formaline, dehydrated, mounted in paraffin, sectioned, and stained with hematoxylin and eosin (HE). T-bet and GATA-3 mRNA were detected in the lung sections using an in situ hybridization system according to the manufacturer’s instructions. The hybridization signal of T-bet mRNA was detected by the streptavidin-biotin-peroxidase complex (SABC-POD) method with the aminoethyl carbazole (AEC) substrate system, and the GATA-3 mRNA was visualized by the streptavidin-biotin-alkaline phosphatase complex (SABC-AP) technique with the diaminobenzidine (DAB) substrate system[16].
Western blot analysis for T-bet and GATA-3 Eighty micrograms of the right lung tissue of 1 rat was frozen immediately and stored in liquid nitrogen until required. The lungs were then homogenized using an electrical homogenization machine, and the proteins were extracted in phosphate-buffered saline (PBS) in the presence of protease inhibitors (6.75% aprotinin, 312 µg/mL trypsin inhibitor) and 0.62% NP-40. Protein concentrations were determined by a protein assay according to the manufacturer’s instructions (Bio-Rad Laboratories Inc, Hercules, CA, USA). The standard curve was performed using 0, 4, 8, and 16 µg of BSA per mL.
The total proteins of the 50 μg isolated from the right lung was separated by 15% SDS-PAGE and blotted onto a nitrocellulose membrane. Equal loading was assessed with Ponceau’s solution (Sigma, St Louis, Missouri, USA). The membrane was then incubated in blocking solution (5% dry milk in PBS) for 1 h at room temperature and subsequently exposed to 0.8 µg/mL of antibodies, anti-T-bet and anti-GATA-3, overnight at 4 ºC. The day after, the membrane was incubated with peroxidase-conjugated anti-rat IgG (1:2500) for 60 min at room temperature. The density of T-bet or GATA-3 stained bands relative to beta-actin expression was quantified with Quantity ONE densitometry software (PDI Imageware Systems, Huntington Station, NY, USA).
Statistical analysis All data were expressed as mean± standard deviation (SD), and the difference was analyzed with nonparametric ANOVA. Statistical analysis was performed using SPSS software (Version 11.5 for Windows, Chicago, IL, USA). The acceptable level of statistical significance was set at a probability of less than 0.05.
Results
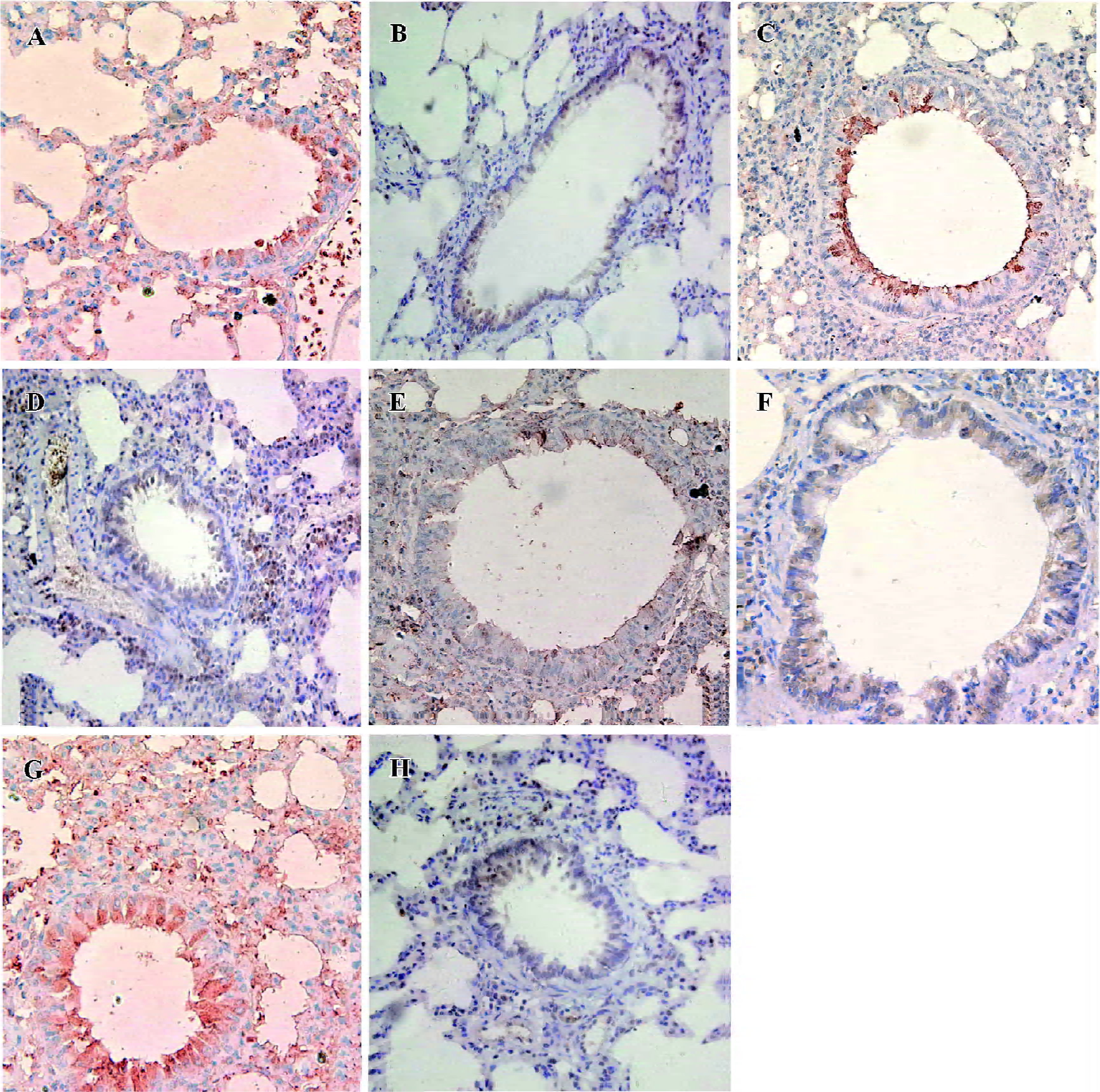
Lung histology and inflammatory cell infiltration in BALF The lungs of the OVA-sensitized and antisense animals showed significantly patchy inflammatory cells infiltration that was located predominantly around the airways compared with the saline-sensitized and nonsense animals (Figure 1). The total cellularity of BALF recovered from the rats in the nonsense group was not significantly different from the saline animals (P>0.05), but differential cell count revealed a significantly increased percentage of eosinophils in both the antisense and OVA-exposed animals (3.0%±1.0% and 4.5%±0.9%, respectively) compared with the saline animals (0.6%±0.1%, both P<0.01) (Table 2).
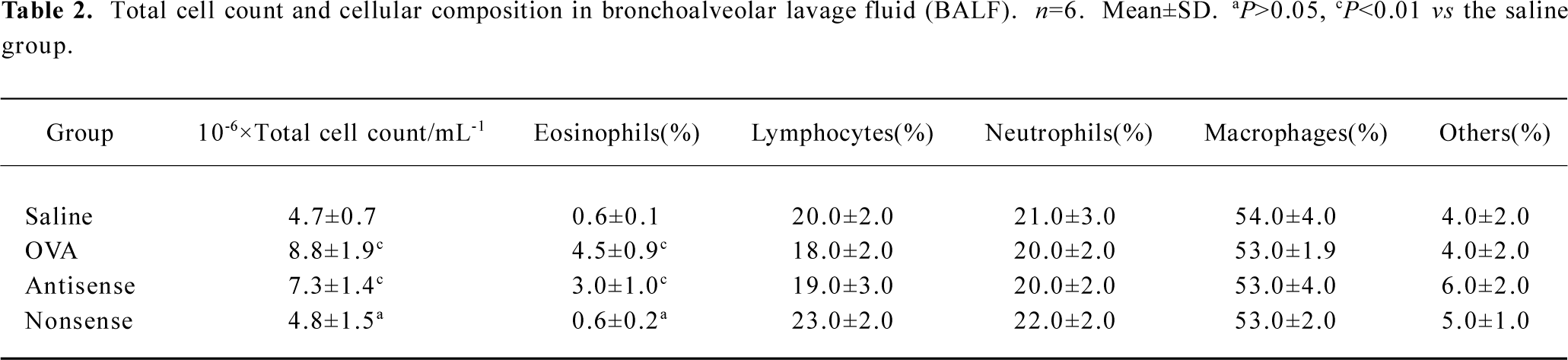
Full table
Levels of IL-4, IL-5, and IFN-γ in BALF The levels of IL-4 and IL-5 in the BALF of both the antisense group (95.2±21.7 µg/L and 19.5±4.9 µg/L, respectively) and the OVA-sensitized group (104.6±18.3 µg/L and 23.0±5.6 µg/L, respectively) were significantly higher than those in the nonsense group (41.6±18.2µg/L and 4.3±1.8 µg/L, respectively) and the saline group (39.2±19.9 µg/L and 3.1±1.4 µg/L, respectively; all P<0.01), but the level of IFN-γ in both the antisense (288.2±18.9 µg/L) and OVA-sensitized (261.5±30.7 µg/L) animals was lower compared with the nonsense (346.8±19.8 µg/L) and the saline (352.5±22.2 µg/L) animals (all P<0.01; Figure 2).
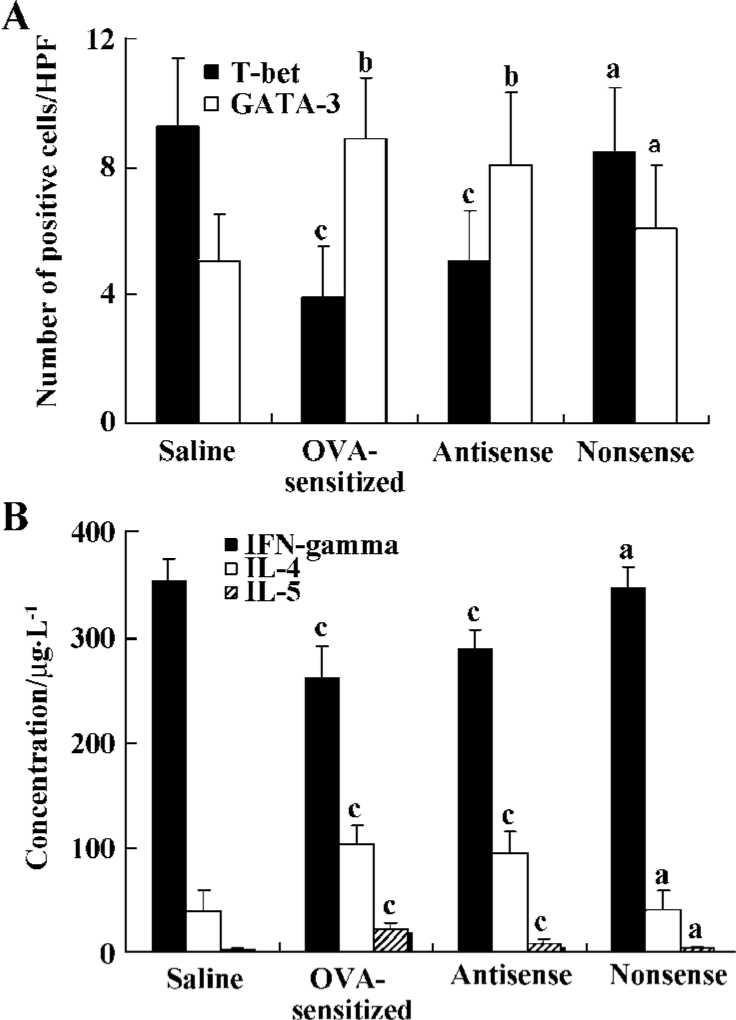
In situ hybridization detection for T-bet and GATA-3 mRNA T-bet mRNA-positive cells exhibited red staining, and the GATA-3 mRNA positive cells had brown staining in the cytoplasma. The number of T-bet mRNA positive cells per high-power field (HPF) in both the antisense group (5.1±1.6 cells/HPF) and the OVA-sensitized group (3.9±1.6 cells/HPF) was significantly lower compared to the nonsense group (8.5±2.1 cells/HPF) and saline group (9.3±2.1 cells/HPF; P<0.01). Whereas the number of GATA-3 mRNA positive cells per HPF in the antisense group (8.1±2.3 cells/HPF) and the OVA-sensitized group (8.9±1.9 cells/HPF) was significantly higher compared with that in the other groups (6.1±2.0 cells/HPF and 5.1±1.5 cells/HPF, respectively; P<0.05; Figures 1,2).
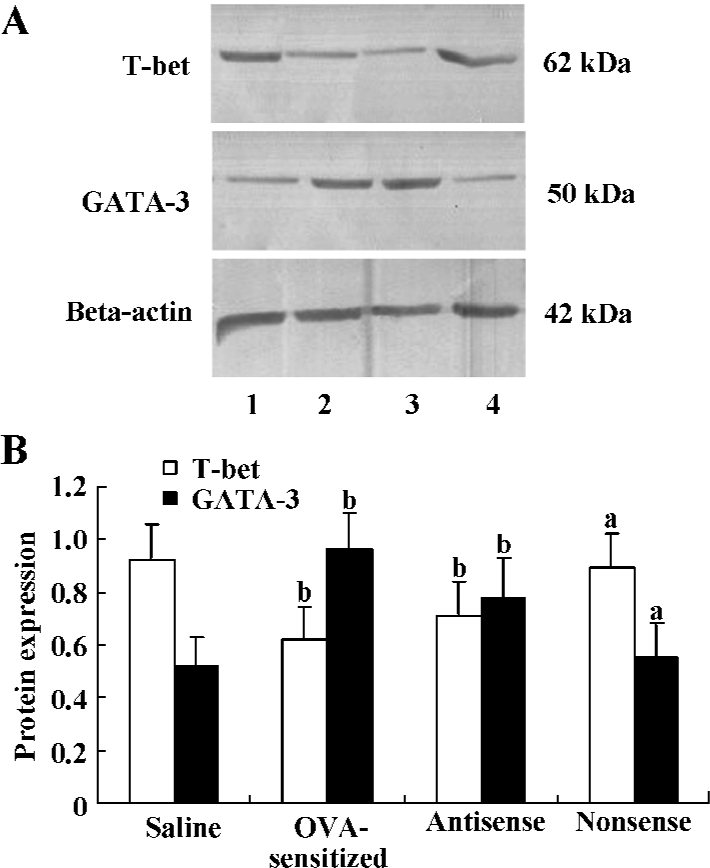
Expression of T-bet and GATA-3 protein Western blot analysis of 50 μg total protein derived from the right lung was performed in order to assess the local T-bet and GATA-3 expression. Down-regulation of T-bet relative to beta-actin protein expression was observed in the antisense T-bet-treated (0.71±0.13) and OVA-sensitized (0.62±0.12) lungs, but not in the lungs treated with nonsense oligonucleotides (0.89±0.13) compared with the saline group (0.92±0.14; all P<0.05). On the contrary, protein expression of GATA-3 relative to beta-actin was up-regulated in the antisense (0.78±0.15) and OVA-sensitized (0.96±0.14) groups compared with the saline group (0.52±0.11; all P<0.05; Figure 3).
Discussion
In recently published studies, mice with a targeted deletion of the T-bet gene and severe combined immunodeficient mice receiving CD4+ cells from T-bet knock-out mice spontaneously demonstrated multiple physiological and inflammatory features characteristic of asthma, indicating that the lack of T-bet gene plays a crucial role in the development of Th2-mediated lung inflammation[14]. However, it remains unclear whether local blockade of T-bet expression in the lung would lead to airway inflammation.
AS-ODNs are now commonly used as a selective strategy to inhibit the expression of a variety of genes[17]. The lung represents an ideal organ to use AS-ODN, given that its large surface area and the surfactant that could facilitate the cellular absorption of ODN. Aerosol of intratracheal administration could avoid the potential toxicity of systemic administration. Therefore, antisense technique and AS-ODN intratracheal administration were applied in the present study. After the intratracheal delivery of T-bet AS-ODN, but not T-bet nonsense-ODNs, T-bet mRNA positive cells and the level of T-bet protein markedly decreased in the lungs.
T-bet can induce over-expression of IFN-γ[8]. Upon IFN-γ stimulation, janus kinase (Jak) 1 and Jak 2 become tyrosine phosphorylated and activated, and induce tyrosine phosphorylation and homodimerization of the transcriptional activator signal transducer and activator of transcription protein (STAT)-1[9–11]. Subsequently, the Stat1:Stat1 homodimer complex translocates to the nucleus of the cells where it binds to IFN-activated site elements present in the promoters of regulated genes to initiate transcription. It has been recently reported that activated STAT-1 could induce T-bet. T-bet then induces IFN-γ itself in a positive feedback loop, promoting the stabilization of the Th1 phenotype[8,18]. In the present study, we demonstrate that T-bet AS-ODN significantly reduce the level of IFN-γ in the BALF which may be induced by local blockade of T-bet expression[19].
Cell lineage specification depends on both gene activation and gene silencing, and in the differentiation of T-helper progenitors to Th1 or Th2 effector cells, this requires the action of 2 opposing transcription factors: T-bet and GATA-3. Imbalanced T-cell-specific transcription factors T-bet and GATA-3 can contribute to type 2 T-helper cell polarization in asthmatic pathogenesis[12,20,21]. GATA-3 could intrinsically suppress T-bet[22] and T-bet can in turn repress the Th2 lineage through the tyrosine kinase-mediated interaction between the 2 transcription factors that interferes with the binding of GATA-3 to its target DNA[23]. Therefore, decreased expression of T-bet by local blockade with AS-ODN could lead to the up-regulation of GATA-3 and the imbalance of the T-bet and GATA-3 ratio in the lungs.
GATA-3 is necessary and sufficient for IL-4 production[24] and the regulatory elements outside of 800-bp proximal promoter are required to direct full IL-4 gene activity[25]. Some published studies have shown the binding of GATA-3 to 3 sites in the human IL-5 promoter[26] where GATA-3 acts directly to induce IL-5 promoter activity[5]. IL-5 is a key regulator of growth and differentiation of eosinophils. In this study, the levels of IL-4 and IL-5 in BALF were further increased, and eosinophilia, a hallmark of asthma, became apparent after 28 d administration of AS-ODN in antisense rats. Our data demonstrates that targeted inhibition of T-bet expression in the local lung by an AS-ODN, in the absence of OVA, results in a selective alteration in the pattern of cytokine expression in the BALF and eosinophilia in airways similar to those in the OVA-sensitized rats.
The Th1/Th2 concept is needed in order to understand the pathophysiology of certain diseases. The present study implies that a novel approach of the local delivery of retroviral gene transduction of T-bet or administration of a T-bet-specific AS-ODN or small interfering RNA (siRNA) has the potential advantage of increasing or suppressing the expression of various Th1 cytokines simultaneously rather than increasing or suppressing the activity of a single cytokine. Several hazards and ethics associated with gene transfer have been verified by clinical trials[27-30]. Interestingly, recently published studies have found that traditional medicines, such as astragalus[31], several protopanaxatriol-type ginsenosides[32], and Chung-Yeul-Gue-Soup-Sa-Gan-Tang[33], could enhance T-bet gene activity and modulate Th1/Th2 lineage development. Therefore, some Chinese medicinal herbs could be a complementary approach in enhancing T-bet activity by regulating the skewed balance of Th1/Th2 in treating asthma.
In conclusion, our results show that antisense-induced local blockade of T-bet expression leads to airway inflammation with a selective alteration in patterns of cytokine expression and eosinophilia similar to those in mice lacking T-bet. Furthermore, the data support the importance of T-bet as a key role in the pathogenesis of asthma, and suggest that T-bet would provide possible therapeutic benefits for asthma sufferers.
Acknowledgements
The authors wish to thank Ms Dan XU of the Department of Respiratory Laboratory, Dr Bing LIU and Dr Zhen-yu DING of the Key Laboratory of Biotherapy of Human Diseases of Ministry of Education for their technical help with in situ hybridization detection and Western blot analysis. Moreover, the authors greatly acknowledge Jon P COLIAS of San Francisco State University and Ze-yu XIONG of the Baylor College of Medicine for helpful assistance with editing this manuscript.
References
- Wang ZL. Research on airway inflammation: present status in mainland China. Chin Med J (Engl) 2005;118:1007-14.
- Kon OM, Kay AB. T cells and chronic asthma. Int Arch Allergy Immunol 1999;118:133-5.
- Holgate ST. The epidemic of allergy and asthma. Nature 1999;402 Suppl:B2-B4.
- Ray A, Cohn L. Th2 cells and GATA-3 in asthma: new insights into the regulation of airway inflammation. J Clin Invest 1999;104:985-93.
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem 1997;272:21597-603.
- Lee HJ, O’Garra A, Arai K, Arai N. Characterization of cis-regulatory elements and nuclear factors conferring Th2-specific expression of the IL-5 gene: a role for a GATA-binding protein. J Immunol 1998;160:2343-52.
- Finottto S, De Sanctis GT, Lehr HA, Herz U, Buerke M, Schipp M, et al. Treatment of allergic airway inflammation and hyperresponsiveness by antisense-induced local blockade of GATA-3 expression. J Exp Med 2001;193:1247-60.
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000;100:655-69.
- Zhang Y, Apilado R, Coleman J, Ben-Sasson S, Tsang S, Hu-li J, et al. Interferon gamma stabilizes the T helper cell type 1 phenotype. J Exp Med 2001;194:165-72.
- Hardy KJ, Manger B, Newton M, Stobo JD. Molecular events involved in regulating human interferon-gamma gene expression during T cell activation. J Immunol 1987;138:2353-8.
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol 2003;21:713-58.
- Liu CT, Wang G, Luo FM, Wang ZL, Liu R, Wang CL. Imbalanced T cell-specific transcription factors T-bet and GATA-3 contributes to types 2 T helper cell polarization in asthmatic patients. Zhonghua Jie He He Hu Xi Za Zhi 2004;27:398-402.
- Raby BA, Hwang ES, Steen KV, Tantisira K, Peng S, Litonjua A, et al. T-bet polymorphisms are associated with asthma and airway hyperresponsiveness. Am J Respir Crit Care Med 2006;173:64-70.
- Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FH, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science 2002;295:336-8.
- Wu ZQ, Xu YP, Xiang H, Shen HH. Effects of CpG oligodeo-xynucleotide on transcription factors GATA-3 and T-bet mRNA expression in asthmatic mice. Acta Pharmacol Sin 2005;26:1117-22.
- Mao H, Yin KS, Wang ZL, Li FY, Zhang XL, Liu CT, et al. Effects of glucocorticoid and cysteinyl leukotriene 1 receptor antagonist on CD34+ hematopoietic cells in bone marrow of asthmatic mice. Chin Med J (Engl) 2004;117:592-7.
- Crooke ST. Molecular mechanisms of action of antisense drugs. Biochim Biophys Acta 1999;1489:31-44.
- Paul WE A, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA 2001;98:15137-42.
- Lovett-Racke AE, Rocchini AE, Choy J, Northrop SC, Hussain RZ, Ratts RB, et al. Silencing T-bet defines a critical role in the differentiation of autoreactive T lymphotytes. Immunity 2004;21:719-31.
- Yates A, Callard R, Stark J. Combining cytokine signalling with T-bet and GATA-3 regulation in Th1 and Th2 differentiation: a model for cellular decision-making. J Theor Biol 2004;231:181-96.
- Ritz SA, Cundall MJ, Gajewska BU, Swirski FK, Wiley RE, Alvarez D, et al. The lung cytokine microenvironment influences molecular events in the lymph nodes during Th1 and Th2 respiratory mucosal sensitization to antigen in vivo. Clin Exp Immunol 2004;138:213-20.
- Grogan JL, Locksley RM. T helper cell differentiation: on again, off again. Curr Opin Immunol 2002;14:366-72.
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 2005;307:430-3.
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997;89:587-96.
- Wenner CA, Szabo SJ, Murphy KM. Identification of IL-4 promoter elements conferring Th2-restricted expression during T helper cell subset development. J Immunol 1997;158:765-73.
- Schwenger GT, Fournier R, Kok CC, Mordvinov VA, Yeoman D, Sanderson CJ. GATA-3 has dual regulatory functions in human interleukin-5 transcription. J Biol Chem 2001;276:48502-9.
- Gao Z, Kang Y, Xu Y, Shang Y, Gai J, He Q. Inhibition of allergic responsiveness in a murine asthma model via IFN-gamma transgene expression. Chin Med J (Engl) 2002;115:1470-4.
- Weigt H, Nassenstein C, Tschemig T, Muhlradt PF, Krug N, Braun A. Efficacy of macrophage-activating lipopeptid-2 combined with interferon-gamma in a murine asthma model. Am J Respir Crit Care Med 2005;172:566-72.
- Kimmelman J. Recent developments in gene transfer: risk and ethics. BMJ 2005;330:79-82.
- Couzin J, Kaiser J. Gene therapy as Gelsinger case ends, gene therapy suffers another blow. Science 2005;307:1028.
- Wei H, Sun R, Xiao W, Feng J, Zhen C, Xu X, et al. Traditional Chinese medicine Astragalus reverses predominance of Th2 cytokines and their up-stream transcript factors in lung cancer patients. Oncol Rep 2003;10:1507-12.
- Yu JL, Dou DQ, Chen XH, Yang HZ, Guo N, Cheng GF. Protopanaxatriol-type ginsenosides differentially modulate type 1 and type 2 cytokines production from murine splenocytes. Planta Med 2005;71:202-7.
- Ko E, Park JW, Rho S, Cho C, Park S, Ko S, et al. Chung-Yeul-Gue-Soup-Sa-Gan-Tang, traditional Korean medicine, enhances CD4(+) T cell, activates and modulates Th1/Th2 lineage development. J Pharmacol Sci 2004;94:359-67.
