Identification and classification of all potential hemolysin encoding genes and their products from Leptospira interrogans serogroup Icterohae-morrhagiae serovar Lai1
Introduction
Leptospirosis is a worldwide zoonosis caused by pathogenic species of Leptospira, particularly, Leptospira interrogans[1]. Pathogenic Leptospira infection causes leptospirosis (Weil’s syndrome), which manifests with jaundice and renal failure[2–4] along with prominent respiratory symptoms. In some cases, severe pulmonary hemorrhages may happen, which may lead to sudden death. Although potential virulence factors, such as hemolysin, lipopolysac-chride (LPS), and heat shock proteins, are suggested for leptospiral infection, the pathogenetic mechanism of leptos-pirosis is yet to be clarified.
Among these suggested virulence factors, bacterial hemolysin has been demonstrated in several pathogenic bacteria[5,6] and a limited number of them have been identified in L interrogans[7]. Annotation of the complete genomic sequence of L interrogans serogroup Icterohaemorrhagiae serovar Lai[8] indicated that there were ten putative hemolysin genes located on the large chromosome (CI, GB: AE010300), of which, one (LA3540) had been previously identified[7]. Our current work characterized all the other genes, except LA0177 because of its extremely short nucleotide sequence. The classification of the hemolytic activities and their expression and secretion in L interrogans were investi-gated.
Materials and methods
Bacterial strains and plasmids The virulent L interro-gans serogroup Icterohaemorrhagiae serovar Lai type strain 56601 used in this study was maintained by the Institute for Infectious Disease Control and Prevention, Beijing, China. The avirulent strain of L interrogans serogroup Ictero-haemorrhagiae serovar Lai (strain IPAV) was given as a gift by Dr PICARDEAU, M (Institute Pasteur, Paris, France). Strains were grown in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium[1] at 28 °C under aerobic conditions and collected at a density of approximately 1×108 bacteria per mL. Escherichia coli DH5α and BL21 (DH3) were used for the cloning and expression of hemolysin candidates, respectively. The pUCm-T and pET28b plasmids were served as the vectors for cloning and expression, respectively.
Characterization of hemolysin candidates First, coding sequences (CDSs) potentially encoding hemolysin candidates were identified during the genomic annotation[8]. Second, amino acid sequences of those hemolysin candidates were analyzed with the SWISS-PROT/TrEMBL non-redundant databases[9] to obtain homologous proteins, which were further compared with BioEdit (Tom Hall, North Carolina State University, Carolina, USA). Third, the domain structures of the hemolysin candidates were predicted by Pfam[10], PROSITE[11] or ProDom[12]. The secondary structures of the proteins were predicted by Jpred2 [13]. Finally, multiple sequence alignments were made for sphingomyelinase-like hemolysin homologous proteins using clustW, and Mega2 (Sudhir Kumar, Arizona State University, Arizona, USA) was used to establish their phylogenetic tree.
Cloning, expression, and purification of the recombinant hemolysin candidates in E coli Genomic DNA was isolated from L interrogans strain 56601 cultivated in EMJH medium. The hemolysin candidate genes were obtained by polymerase chain reaction (PCR), ligated with pUCm-T vector and transformed into DH5α cells. The confirmed recombinant plasmids were digested with corresponding enzymes, ligated with pET28b vector and then transformed into BL21 (DH3) cells. E coli cells were grown in Luria-Bertani medium supplemented with kanamycin at 50 mg/L at 37 °C. Protein expression was induced at A600 of 0.6 by addition of isopropyl-beta-D-thiogalactopyranoside (IPTG) at 0.6 mmol/L for 3 h. The harvested cells were suspended in Tris-HCl buffer (20 mmol/L Tris-HCl, pH 7.9) and lysed by sonication. The insoluble inclusion bodies were dissolved in the buffer (Tris-HCl 20 mmol/L, pH 7.9, NaCl 0.5 mol/L, 10% glycerol, guanidium HCl 6 mol/L) and then centrifuged at 20 000×g for 20 min. The soluble supernatant was applied to Ni-NTA His-Bind resin and the His-tag fusion protein was eluted by an imidazole gradient from 10 mmol/L to 1000 mmol/L in the elution buffer (20 mmol/L Tris-HCl pH 7.9, 0.5 mol/L NaCl, 10% glycerin). The purified proteins were analyzed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) with coomassie brilliant stain.
Hemolytic activity assay on sheep blood agar plates Sheep blood plates, 10% (v/v) supplemented with 25 mmol/L MgCl2 and 100 mg/L kanamycin, were used to measure hemolytic activities. Lysate of E coli cells expressing hemolysin candidate proteins (100g/L) was dropped onto the plates, and the plates were incubated at 37 °C for 16 h, and then placed at 4 °C for 30 min. Sphingomyelinase C (0.1 U, Sigma, St Louis, MO, USA) and lysate of E coli cells harboring pET28b (100 g/L) were used as positive and negative controls, respectively.
Sphingomyelinase assay by thin-layer chromatography and HPLC The biphasic system consisted of an ether:methanol (9:1 v/v) organic phase containing sphingomyelin (2 g/L) and a water phase (25 mmol/L MgCl2) containing the lysate of E coli cells expressing hemolysin candidate proteins (100 g/L). This biphasic solution was shaken at 37 °C for 4 h. After that, the organic phase (15 μL) was applied on a silica gel-60 coated glass plate. The chromatogram was developed with a mobile phase (chloroform:methanol:water:25% ammonia 58.0:35.0:3.5:3.5 v/v). Lipids were visualized by spraying on a plate with 30% H2SO4 at 110 °C for 10 min. For HPLC, sphingomyelinase activity was determined in a biphasic system as described above, except for the organic phase containing sphingomyelin (1 g/L). The organic phase (7.5 μL) was applied to HPLC (YWG C18 5 μm, 200 mm×4.6 mm, China) and the elutes were monitored by absorption at 207 nm. The mobile phase was a mixture of acetonitrile:methanol:water (154:45:81 v/v). The flow rate was 1 mL/min. Sphingomyelinase C (0.1 U) and lysate of E coli cells harboring pET28b (100 g/L) were used as positive and negative controls, respectively.
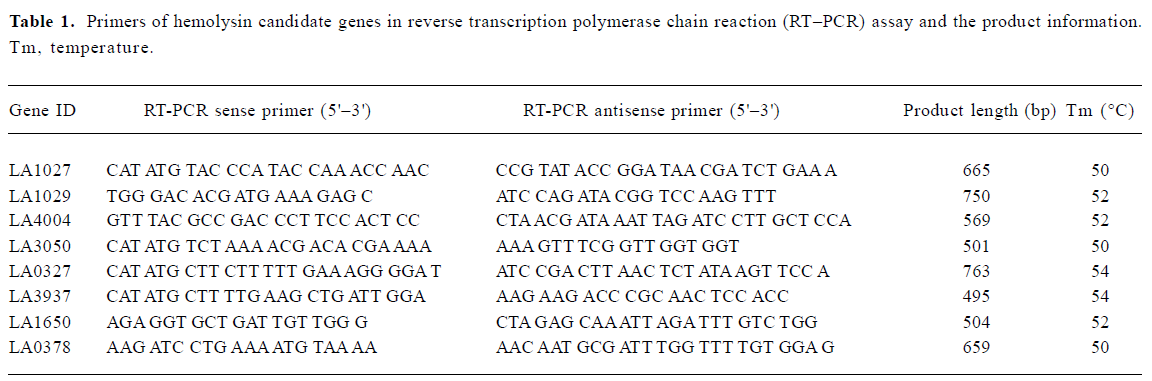
Validation of hemolysin encoding gene expressions in Leptospira interrogans strain Lai by reverse transcription RCR and Western blot According to the manufacturer’s instructions, total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, California, USA) from L interrogans strain Lai cultivated in EMJH medium. Total RNA (1 μg) from each sample was reverse-transcribed into cDNA according to the instructions provided with the cDNA Synthesis Kit (Invitrogen, Carlsbad, California, USA). Equal amounts of the product of the reverse transcription reaction were subjected to PCR amplification. The primers and related information are shown in Table 1. After amplification, 5 μL of each PCR reaction product was electrophoresed on a 1.5% (w/v) agarose gel containing ethidium bromide (0.5 mg/L). For Western blot analysis, L interrogans strains cultivated in EMJH medium were harvested by centrifugation at 14 600×g for 10 min, electrophoresed on 10% SDS-PAGE gels and electrotransferred to nitrocellulose membrane under a constant voltage of 5 mA/cm2 for 1 h. The blot was first masked by the blotting buffer (10% skim milk in 10 mmol/L Tris-HCl buffer with NaCl 150 mmol/L and 0.1% Tween-20) for 2 h and then incubated with rabbit anti-hemolysin antibody (1:5000) for 2 h. After being washed with TBST 3 times, the blot was incubated for 1 h with alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG antibody (1:7500) and the color was developed by addition of bromo-chloro-indoryl phosphate/nitroblue tetrazolium (BCIP/NBT) for AP reaction.
Full table
Detection of hemolysin secretion in L interrogans Briefly, the culture supernatant (100 ng/well) of L interrogans in Korthof[1] was coated onto 96-well ELISA plates by incubation at 37 °C for 2 h. After the removal of blocking solution and washing 3–5 times with phosphate-buffered saline buffer (PBS) containing 0.1% Tween (PBS-Tween), anti-hemolysin antibodies were incubated in the plate at 37 °C for 2 h. The plate was washed 3–5 times with PBS-Tween and then incubated with AP-conjugated goat anti-rabbit IgG antibody (1:1500) at 37 °C for 1 h. The plate was washed 3–5 times with PBS-Tween and incubated with the BCIP/NBT for 20 min in the dark. Absorbance at 492 nm was recorded using an automated ELISA microplate reader.
Results
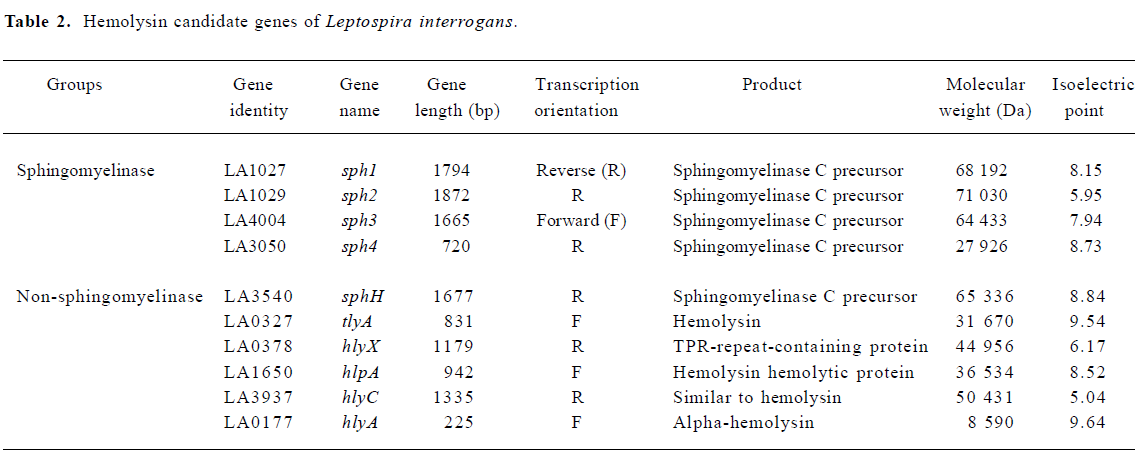
The in silico structural analysis indicated that there were at least two kinds of hemolysin candidates from L interrogans Genomic annotation of L interrogans strain Lai indicated that there were 10 genes putatively encoding proteins highly similar to hemolysins reported in the NCBI/Genbank and SwissProt/TrEMBL databases (Table 2).
Full table
Domain structure analysis of the candidate proteins indicated that 5 of them had barring of the characteristic domains of the phosphatase family. Specifically, the Pfam-based domain analysis indicated that all of LA1027, LA1029, LA4004, and LA3050 barred a conserved domain PF03372 of the phosphatase family. The ProDom-based analysis showed that all of the proteins, except LA3050, were highly similar to L interrogans serovar hardjo sphingomyelinase-like hemolysin (SP17627/SwissProt), barring phosphatase domains of PD011673, PD447657, and PD041204 in similar regions of the proteins. Although LA3050 was reasonably similar to SP17627 (67%) and also had the PD041204 domain, it lacks the other two domains mainly because of its short primary peptide sequence. Although LA3540 had PD011673, PD041204, and the phosphatase conserved domain PF03372, LA3540 had previously been identified as a pore-forming hemolysin[5].
Amino acid sequence analysis of these sphingomye-linase-like hemolysin candidates indicated that the conserved Mg2+-complexing glutamic acid and asparagine involved in substrate binding were identified in LA1029, Glu200, and Asn343, corresponding to the Glu131 and Asn274 of SP17627, respectively. For LA4004, only Asn267 was identified as corresponding to the conserved Asn274 of SP17627. Protein secondary structures were predicted by Jpred2, and their similarities are shown in Figure 1.
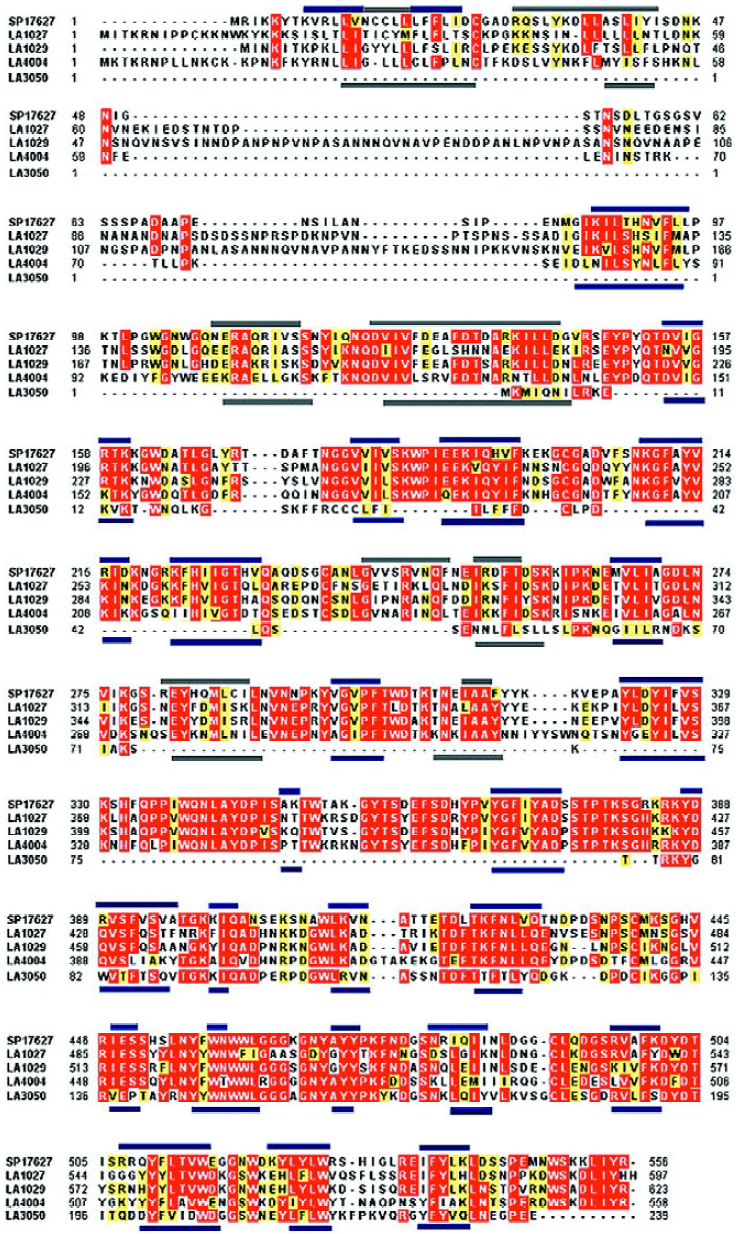
The remaining 5 candidate proteins have little sequence similarity to those of sphingomyelinase-hemolysin proteins. Based on the in silico analysis, they do not have any phosphatase family domains or share any common domains. Therefore, these hemolysin candidates, if they are active, may function based on the mechanism of pore formation and other mechanisms.
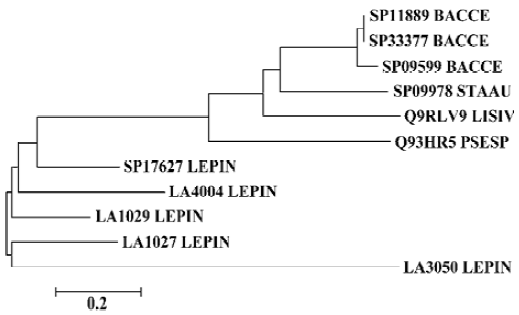
Phylogenetic analysis was performed on the sphingo-myelinase-hemolysin proteins. An neighbor-joining unrooted phylogenetic tree was established including sequences of SP17627, LA1027, LA1029, LA4004, LA3050, and other similar proteins, with a cut-off amino acid identity of 53% and similarity of 67%. Hemolysins from Staphylococcus aureus, Bacillus cereus, Listeria ivanovii, and Pseudomonas sp strain TK4 made up one branch of the tree, and the sphingo-myelinase-like proteins of L interrogans made up another branch (Figure 2).
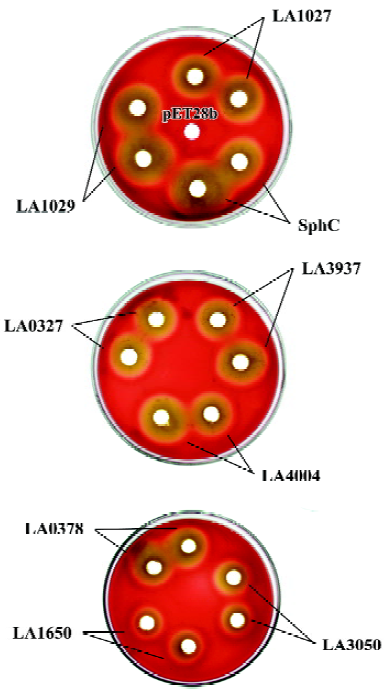
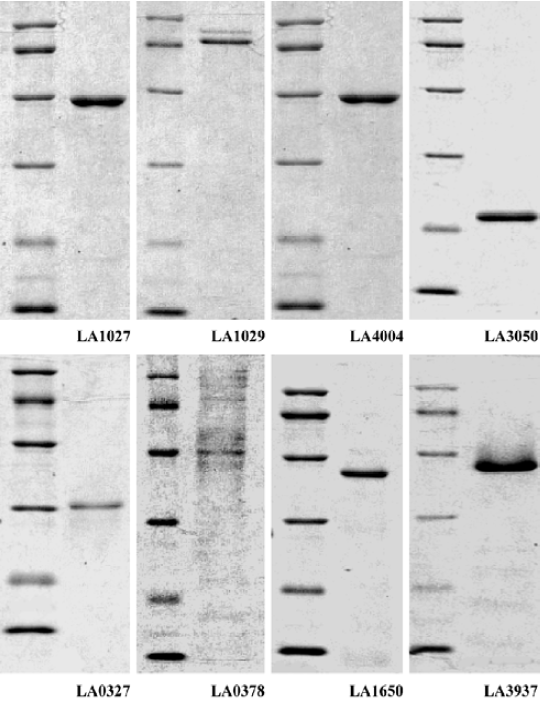
The hemolytic activities of the recombinant hemolysin candidates from L interrogans were determined Among the 10 hemolysin candidates from L interrogans, LA3540 had previously been identified[7], whereas LA0177 is extremely short in its sequence and has no detectable phosphatase domain. Therefore, only the remaining 8 hemolysin candidate genes were cloned into E coli and the recombinant proteins were purified to homogeneity determined by SDS-PAGE followed by coomassie brilliant stain (Figure 3).
Crude cell lysates of E coli expressing cloned hemolysin candidates were spotted onto sheep blood agar plates and, after incubation at 37 oC for 16 h, clear hemolytic zones were observed for all the candidates via a cold-warm hemolytic procedure (Figure 4). The hemolytic zone of LA1029 appeared at first and the area of hemolysis was the biggest. Clear hemolytic zones of LA1027 and LA4004 appeared a little later. Hemolysis caused by LA3050 appeared last and the area of the hemolytic zone was the smallest. For those predicted to be in the non-sphingomyelinase-hemolysin family, hemolytic abilities were also different: LA3937 was the highest and LA0327, LA0378, and LA1650 were second highest, third highest, and lowest, respectively.
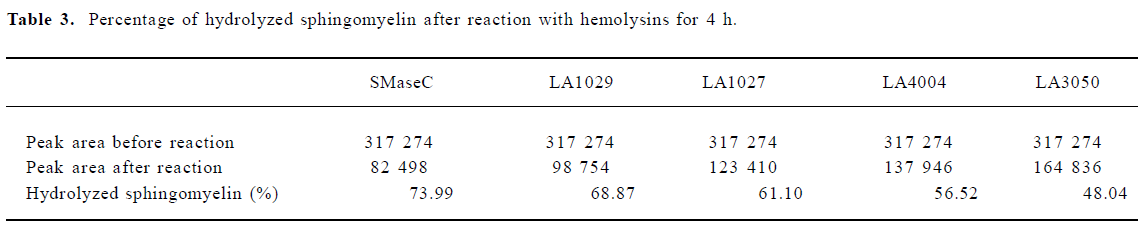
The sphingomyelinase activities of hemolysin candidates were detected The sphingomyelinase activities of recombinant hemolysin candidates were detected by TLC assay. Results showed that only LA1027, LA1029, LA4004, and LA3050 could hydrolyze sphingomyelin. These results were confirmed by HPLC assay. The peak area of sphingomyelin was diminished after hemolysin treatment and the percentages of hydrolyzed sphingomyelin are displayed in Table 3. The percentages of hydrolyzed sphingomyelin of sphingo-myelinase (SphC), LA1029, LA1027, LA4004, and LA3050 were 73.99%, 68.87%, 61.10%, 56.52%, and 48.04%, respectively. LA3937, LA0327, LA0378, and LA1650 could not hydrolyze sphingomyelin under the same condition.
Full table
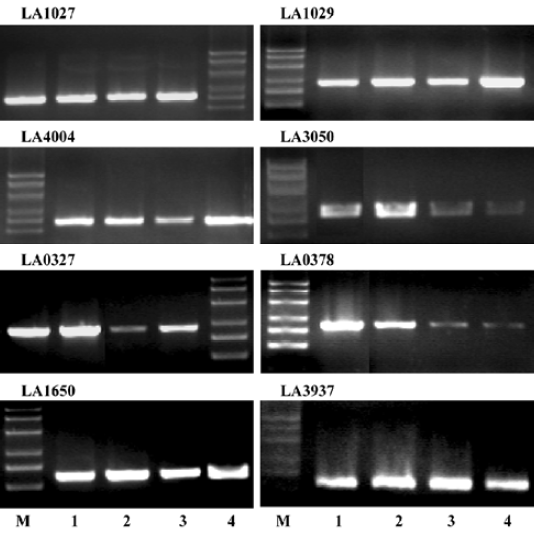
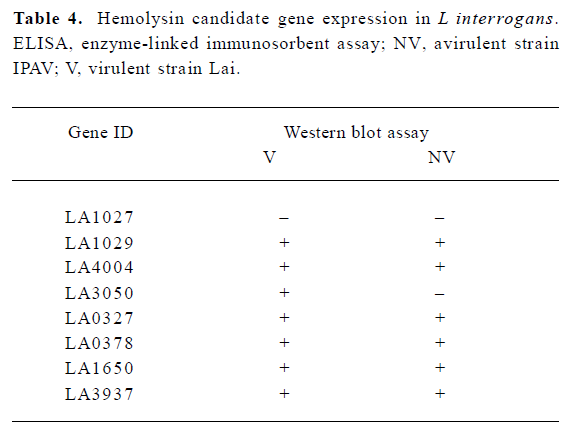
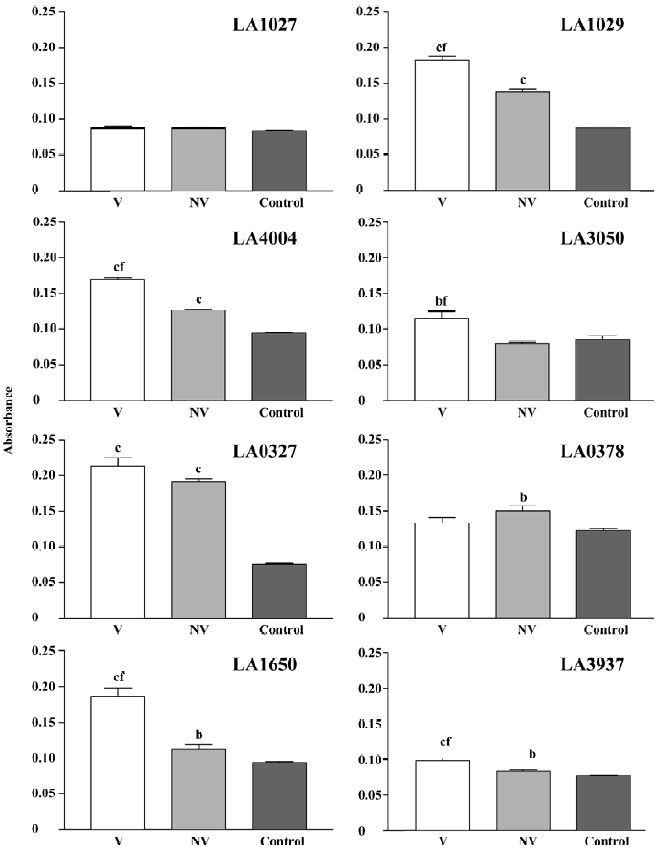
Hemolysin gene expression and hemolysin secretion in L interrogans RT-PCR analysis indicated that all the 8 candidates, LA0327, LA0378, LA1027, LA1029, LA1650, LA3050, LA3937, and LA4004, were transcribed in L interrogans strain Lai and strain IPAV cultivated in EMJH or Korthof culture medium (Figure 5). The transcription levels of some hemolysin encoding genes, LA0378, LA0327, and LA3050, were higher in L interrogans cultivated in Korthof medium than those in the EMJH medium. Western blot can detect hemolysin proteins in strain Lai cell crude extracts for all the candidates except LA1027 (Table 4). In addition, we failed to detect this protein by Western blot in the strain IPAV cultured in EMJH medium. In addition, we failed to detect LA3050 under the same condition, whereas the other 6 hemolysin candidates were all detected. LA1029, LA4004, LA3050, LA1650, and LA3937 were secreted into the environment, as determined by ELISA, and the secretion levels were higher in strain Lai than those in strain IPAV (Figure 6). LA0327 was secreted into the environment both in strain Lai and in strain IPAV. LA1027 was not secreted either in strain Lai or in strain IPAV.
Full table
Discussion
In 1956, Alexander et al[14], for the first time, discovered that L interrogans had a hemolysin-like substance that could hemolyze red blood cells of ruminants, such as sheep, cows, and goats. Its hemolytic activity was completely lost after being heated at 56 °C for 5 min, indicating its heat-labile character. Later, hemolysins were detected in many pathogenic L interrogans[15].
L interrogans gene encoding hemolysin was first cloned from the pathogenic serovar hardjo. This protein obviously possessed both hemolytic and sphingomyelinase C activities[16]. The only hemolysin gene cloned from serovar lai before our efforts was CDS LA3540[7]. Although this gene encoded a hemolytically active protein with a phosphatase-like domain, the purified protein did not have any detectable sphingomyelinase activity. Therefore, it was proposed that it was a transmembrane pore formation protein. The concept of transmembrane pore formation by bacterial protein toxins was first brought forward by Fussle et al[17] to explain the mechanism of action of staphylococcal alpha-toxin and it is apparent today that the majority of medically relevant pathogens produce pore-forming proteins[18]. Many of these toxins have been designated as hemolysins because of their lytic action on red blood cells.
Our studies have proven experimentally that, besides the previously characterized LA3540 and the very short hemolysin candidate gene, LA0177, the recombinant proteins encoded by the candidate genes from L interrogans strain Lai (LA0327, LA0378, LA1027, LA1029, LA1650, LA3050, LA3937, LA4004) were hemolysin. According to the in silico structure analysis, LA1027, LA1029, LA4004, and LA3050 are highly similar to SP17627, the hemolytic sphingomyelinase C from serovar hardjo, with respect to domain organizations, amino acid sequences, and the predicted secondary structures. These predictions were further confirmed by sphingomye-linase assays. Other candidates, LA0327, LA0378, LA1650, and LA3937, were confirmed to be non-sphingomyelinase hemolysins. They are likely either to be pore-forming proteins or to have other mechanisms of hemolytic activity.
Spirochetes are evolutionarily primitive and L interrogans is a facultative free-living pathogen possessing a large number of different kinds of hemolysins. Therefore, questions about the possibilities of convergence, divergence, or other models of evolution should be addressed in the future. In this study, the phylogenetic analysis of sphingomyelinase from L interrogans, both serovar lai and serovar hardjo, with those from other bacteria indicated that the sphingomyelinase hemolysins of L interrogans are closely related but distantly different from those of the others. Thus, this group of hemolysins is more likely to be evolved from a common ancestor with a divergence mechanism. For the other group of hemoly-sins, because of their high level of diversity, the possibility of horizontal gene transfer cannot be excluded.
Almost all of these hemolysins were expressed in vivo under normal culture conditions. Among them, LA1029, LA4004, LA1650, and LA3937 were secreted into the environment and the secretion level was significantly higher (P<0.01) in the virulent strain Lai than in the avirulent strain IPAV (Figure 6). LA0327 was secreted into the environment both in strain Lai and IPAV without significant difference. LA1027 was not expressed or secreted in either strain Lai or IPAV, which was testified by ELISA and Western blot assay (Figure 6, Table 4). The significant difference in hemolysin expression and secretion between the virulent strain Lai and the avirulent strain IPAV may suggest that hemolysins might play an important role in the pathogenesis of Leptospira.
Acknowledgment
We thank Shuang-xi REN for suggestions and comments, and Bao-yu HU and Xiu-gao JIANG for help in bacterial culture preparation.
References
- Faine S. Leptospira and leptospirosis. New York: CRC Press; 1994.
- Kmety E, Dikken H. Classification of the species and history of its serovars. Groningen: University Press; 1993.
- Ramadass P, Jarvis BD, Corner RJ, Penny D, Marshall RB. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol 1992;42:215-9.
- Ozdemir V, Erol E. Leptospirosis in Turkey. Vet Rec 2002;150:248-9.
- Lee SH, Kim S, Park SC, Kim MJ. Cytotoxic activities of Leptospira interrogans hemolysin SphH as a pore-forming protein on mammalian cells. Infect Immun 2002;70:315-22.
- Sakurai J, Honda T, Jinguji Y, Arita M, Miwatani T. Cytotoxic effect of the thermostable direct hemolysin produced by Vibrio parahaemolyticus on FL cells. Infect Immun 1976;13:876-83.
- Lee SH, Kim KA, Park YG, Seong IW, Kim MJ, Lee YJ. Identification and partial characterization of a novel hemolysin from Leptospira interrogans serovar lai. Gene 2000;254:19-28.
- Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole genome sequencing. Nature 2003;422:888-93.
- SWISS-PROT/TrEMBL databases. Europe: Swiss Institute of Bioinformatics. c2004. Available from: and http://www.expasy.org/sprot
- Pfam database. St Louis: Washington University in St Louis. c2004. Available from: http://pfam.wustl.edu/hmmsearch.shtml
- PROSITE database. Europe: Swiss Institute of Bioinformatics. c2004. Available from: http://us.expasy.org/prosite
- ProDom database. Europe: Laboratoire des Interactions Plantes Microorganismes. c2004. Available from: http://prodes.toulouse.inra.fr/prodom/current/html/home.php
- Jpred database. Scotland: University of Dundee. c2004. Available from: http://www.compbio.dundee.ac.uk/~www-jpred/submit.html
- Alexander AD, Smith OH, Hiatt CW, Gleiser CA. Presence of hemolysin in cultures of pathogenic leptospires. Proc Soc Exp Biol Med 1956;91:205-11.
- Bauer DC, Morse EV. Variation and hemolysin production in relation to virulence of Leptospira pomona. Proc Soc Exp Biol Med 1958;98:505-8.
- Cloning of a hemolysin gene from Leptospira interrogans serovar hardjo. Infect Immun 1989;57:2588-90.
- Fussle R, Bhakdi S, Sziegoleit A, Tranum-Jensen J, Kranz T, Wellensiek HJ. On the mechanism of membrane damage by Staphylococcus aureus alpha-toxin. J Cell Biol 1981;91:83-94.
- Bhakdi S, Tranum-Jensen J. Damage to cell membranes by pore-forming bacterial cytolysins. Prog Allergy 1988;40:1-43.