Biodiversity in cultivated Panax notoginseng populations1
Introduction
Panax notoginseng (Burk) F H Chen belongs to Araliaceae. Its root is one of the most highly valued traditional Chinese medical herbs for its hemostatic and restorative properties. In the last 20 years, a great deal of chemical and pharmacological studies on P notoginseng has been carried out. Compounds isolated from notoginseng include saponins, flavonoids, non-protein amino acids, polysaccharide, fatty acids, aliphatic alkenes and peptides. Dammarene-type triterpenoid saponin glycosides are considered to be the primary pharmacologically effective components that exert various effects on the blood, cardiocerebral vascular system, central nervous system and endocrine system[1–6].
According to historical records, P notoginseng has been cultivated for more than 400 years in Wenshan prefecture (Yunnan province, China), the most important notoginseng producing area in China[7]. This ginseng species is unique in the way that it can only be found in farms, not in the wild. There is the question on extent of biodiversity in such cultured ginseng populations. Our previous study on P notoginseng plants from one single farm found variations in both genetic makeup and contents of ginsenosides (Rg1, Rb1, Re, Rd and notoginsenoside R1)[8], suggesting the presence of biodiversity in one cultured population. This research was designed to evaluate biodiversity of P notoginseng species that are represented by random samples collected from farms in 4 major production counties in Wenshan prefecture. The 3-year-old plants were documented for various morphological features and their genetic diversity evaluated by fluorescent amplified fragment length polymorphism (fAFLP) analysis.
Materials and methods
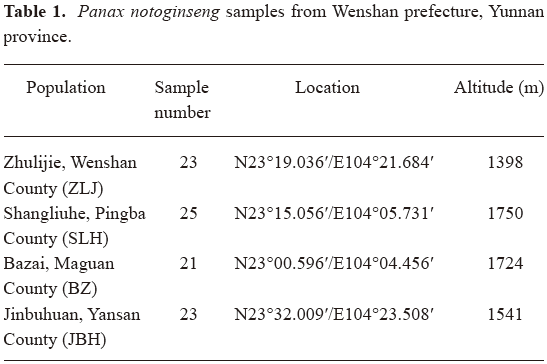
Plant material ninty-two randomly selected 3-year-old P notoginseng plants were harvested in December of 2005 from 4 farms located in 4 counties in Wenshan prefecture (for details see Table 1). Whole plants were photographed and morphological features of plant height (without peduncle), stem color, number of leaves and number of leaflets for each leaf were recorded. One leaf with 8–10 leaflets for each plant was collected and dried with silica gel in sealed plastic bags immediately. Dried leaves were frozen in liquid nitrogen and ground into powder, then stored at –80 ℃. Tap roots were harvested, cleaned, dried in an oven and weighed. For quantitative morphological features, general calculations and Student’s t-test were conducted with Microsoft Excel 2003.
Full table
fAFLP analysis DNA was extracted from powdered leaves using DNeasy Plant Mini kit (Qiagen, CA, USA). The quality of DNA was checked by electrophoresis on 0.8% agarose gel. fAFLP analysis was conducted as previously reported[8]. Selective amplification was carried out with different combinations of EcoRI and MseI selective primers (Applied Biosystems, Foster City, CA, USA). 4 selective AFLP primer combinations were used in this study: EcoRI-ACT/MseI-CAC (1B), EcoRI-ACA/MseI-CAC (2B); EcoRI-ACA/MseI-CTC (2F) and EcoRI-AGG/MseI-CAC (7B). After selective amplification, 0.6 µL PCR product was mixed with ROX size standard (0.6 µL) and Hi-Di formamide (8.8 µL), then subjected to capillary electrophoresis using 3730xl DNA analyzer (Applied Biosystems).
Statistical analysis An AFLP Excel Macro[9] was used to convert allele size data from GeneMapper 3.7 (Applied BiosystemsA) into binary form, to indicate the presence (1) or absence (0) of alleles. GenAlEx 6[10] was used to compute allele frequency in populations, genetic heterozygosity within populations, and the average Nei genetic distance among populations. It was also used for analysis of molecular variance (AMOVA) and principal coordinates analysis (PCA).
Results and Discussion
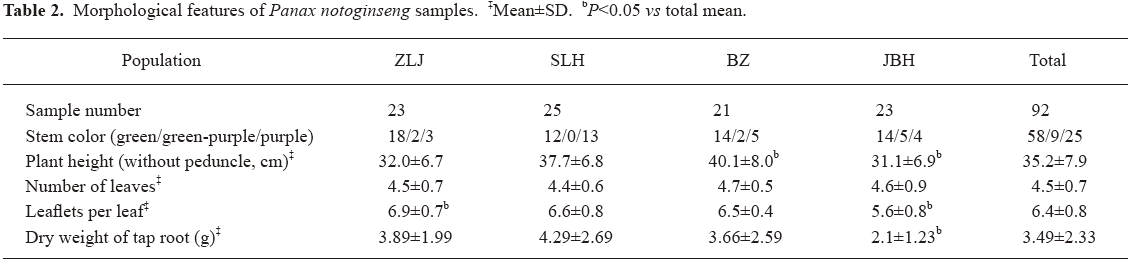
Diversity in morphological features In order to have a good representation of P notoginseng at the species level, samples were collected from the main production area of Wenshan prefecture, Yunnan province of China. Of 4 independent farms located in 4 counties, each had a plantation area of more than 1 hectare and experience of more than 10 years of plantation of P notoginseng (Table 1). Randomly selected 3-year-old plants were harvested from each farm. Morphological features of these plants were recorded (Table 2).
Full table
Differences among samples in morphological features were observed even within the same farms. Plants had different stem colors, were of a wide range of heights, had variable numbers of leaves, had different numbers of leaflets per leaf and, importantly, yielded tap roots of very variable dry weight. It is noted that JBH population had lower plant heights, lower number of leaflets and lower yield of tap root than total population, most likely due to general poor growth of plants on the farm.
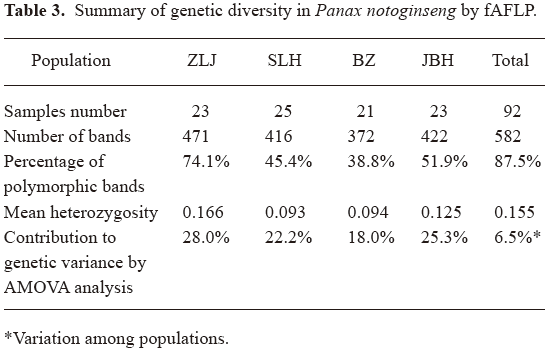
Genetic diversity in P notoginseng fAFLP analysis of 92 samples generated a total number of 582 discrete peaks, among which 509 polymorphic peaks were detected (see Table 3 for a summary).
Full table
In these 4 populations: Zhulijie, Wenshan County (ZLJ), Shangliuhe, Pingba County (SLH), Bazai, Maguan County (BZ), Jinbuhuan, Yansan County (JBH), analyzed by fAFLP, the percentages of polymorphic bands were 74.05%, 45.36%, 38.83% and 51.89%, respectively. Mean genetic heterozygosity were 0.166, 0.093, 0.094 and 0.125. For the total population of 92 samples, the percentage of polymorphic bands was 87.5% and the mean heterozygosity was 0.155. These results were consistent with the genetic diversity reported in wild populations of Panax quinquefolius revealed by random amplified polymorphic DNA (RAPD)[11] and allozyme analysis[12], although results from different techniques are not strictly comparable. Compared with the previous report of genetic diversity in P notoginseng[13], we report similar levels of genetic diversity present in larger numbers of samples from 4 more representing populations. In reference to the highly variable morphological features documented for the same samples, we conclude that a fair level of genetic diversity is present even in cultured P notoginseng species. Further AMOVA attributed most (28.0%+22.2%+18.0%+25.3%=93.5%) genetic diversity to within population variations.
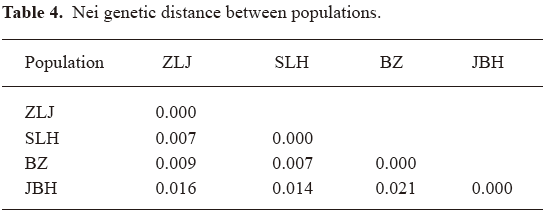
No obvious genetic drift among populations Another objective of this study was to evaluate possible genetic drift or isolation of any population. According to farm owners, these 4 farms had been self sufficient in seeds for many years. Nei genetic distance was used to estimate genetic distance among populations (Table 4).
Full table
It was found that Nei genetic distances between populations were all <0.030, suggesting a lack of obvious genetic diversity between populations. This agrees with the AMOVA result of a small contribution by among population variation to total variation (6.5%). The fact that PCA did not group any population distinctively (data not shown) provides further support to the conclusion that P notoginseng populations are not significantly different from each other genetically. In other words, this species lacks genetic drift or isolation in its populations. This finding might be explained by the lack of geographical isolation and possible exchange of plantation materials during the history of cultivation in Wenshan prefecture. On the other hand, 400 years might not be long for accumulation of significant differences between populations even under full isolation.
The presence of morphology diversity together with genetic diversity in 4 populations of P notoginseng leads to our conclusion that P notoginseng still maintains a fair level of biodiversity at the species level despite a few hundred years of cultivation and the lack of gene exchange with wild plants. We reason that this maintenance of biodiversity in cultivation is associated with the local practice of non-discriminative harvest of seeds in winter time when roots remain underground (the most valuable part, to be harvested in next spring). Another possible contribution factor is the poor germination of seed after storage. Seeds harvested can only be used for next season planting. Poor viability of seeds made it difficult to maintain any unusual plants. The presence of biodiversity in a cultivated population is good for preservation and the future survival of P notoginseng as a species. On the other hand, such diversity is associated with non-uniform performance in farms. There is a lot of room for improvement and this biodiversity will form the basis of future improvement. Individual plants should be evaluated for their morphological traits and pharmacologically active components. One possible approach is to continuously self-pollinate an elite individual to create good performing and more homogeneous populations (in-breeding). An alternative approach is to use 2 genetic divergent parent plants for cross pollination to create hybrids, followed by selection for better performance in hybrid populations (hybrid vigor).
References
- Li SH, Chu Y. Anti-inflammatory effects of total saponins of Panax notoginseng. Acta Pharmacol Sin 1999;20:551-4.
- Jiang KY, Qian ZN. Effects of Panax notoginseng saponins on post hypoxic cell damage of neurons in vitro. Acta Pharmacol Sin 1995;16:399-402.
- Matsuura H, Kasai R, Tanaka O, Saruwatari YI, Fuwa T, Zhou J. Further studies on dammarane-saponins of Sanchi-Ginseng. Chem Pharm Bull 1983;31:2281-7.
- Sengupta S, Toh SA, Sellers LA, Skepper JN, Koolwijk P, Leung HW, et al. Modulating angiogenesis: the yin and the yang in ginseng. Circulation 2004;110:1219-25.
- White CM, Fan C, Chow M. An evaluation of the hemostatic effect of externally applied notoginseng and notoginseng total saponins. J Clin Pharmacol 2000;40:1150-3.
- Yuan J, Guo W, Yang B, Liu P, Wang Q, Yuan H. 116 cases of coronary angina pectoris treated with powder composed of radix ginseng, radix notoginseng and succinum. J Tradit Chin Med 1997;17:14-7.
- Zheng GZ, Yang CR. Biology of and its application. Beijing: Science Press; 1994.
- Hong DYQ, Lau AJ, Yeo CL, Liu XK, Yang CR, Koh HL, et al. Genetic diversity and variation of saponin contents in Panax notoginseng roots from a single farm. J Agr Food Chem 2005;53:8460-7.
- Rinehart TA. AFLP analysis using Genemapper software and an Excel macro that aligns and converts output to binary. Biotechniques 2004;37:186-8.
- Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 2006;6:288-95.
- Schluter C, Punja ZK. Genetic diversity among natural and cultivated field populations and seed lots of American ginseng (Panax quinquefolius L.) in Canada. Int J Plant Sci 2002;163:427-39.
- Cruse-Sanders JM, Hamrick JL. Genetic diversity in harvested and protected populations of wild American ginseng, Panax quinquefolius L (Araliaceae). Am J Bot 2004;91:540-8.
- Zhou SL, Xiong GM, Li ZY, Wen J. Loss of genetic diversity of domesticated Panax notoginseng F H Chen as evidenced by ITS sequence and AFLP polymorphism: A comparative study with P stipuleanatus H T Tsai et K M Feng. J Int Plant Biol 2005;47:107-15.
