Panax ginseng natural populations: their past, current state and perspectives1
Introduction
Panax ginseng C.A. Meyer is a well-known herb of Oriental traditional medicine, used by people for thousands of years as a valuable remedy for various diseases and a tonic drug for the elderly.
In ancient manuscripts (see review in Zhuravlev et al, 1996[1]) ginseng was already described as a rare plant. It was introduced into culture in ancient China and Korea. However, wild-growing ginseng was always considered to be of the most value. It was distributed in the Far Eastern forests on a wide territory, but because of intensive use of wild ginseng roots its area was drastically reduced, particularly in the last century. Indeed, in the first half of XX century wild-growing ginseng plants could be found on the large forestlands of China, Korea and Russian Primorye[2], but nowadays its original distribution area has shrunken to a few habitats in Russia and China. The largest, the Sikhote-Alin population, is located in the southern part of the Sikhote-Alin mountain range. Another population is located in the Nadezhdinsk and Khasan Districts of Primorsky Kray, Russia (Khasan population), and Jilin and Heilongjiang Provinces, China, and a third, the Blue Mountain population, is located in the Spassk District of Primorsky Kray, Russia. Any reports about findings of wild ginseng in China have been rare. In Russia, ginseng was listed in Red Book since 1975 as a threatened species[3]. Nowadays Primorsky Kray of Russia is the only place in the world where natural ginseng populations are still alive and therefore our duty is to conserve this wonderful plant. Ginseng populations are now close to extinction, and some urgent measures have been suggested to protect natural ginseng resources from exhaustion[4]. Moreover, to change the situation and to work out measures for protection of wild-growing ginseng, in 1997 the Primorye regional administration, Regional Committee of Natural Resources and Institute of Biology and Soil Science of the Russian Academy of Sciences (IBSS) elaborated the “Regional Complex Long-term Program of Restoration (Reintroduction) of Primorye’s Ginseng Population up to 2005”. This program was based on many years’ study of various aspects of ginseng biology conducted in IBSS.
During the program execution, IBSS has been providing centers of reintroduction with scientific justification for the selection and propagation of sampled material and for offspring identification, choice of number and emplacement of centers of reintroduction, search for more intact habitats for reintroduction and detailed recommendations for creation of reintroduced populations. During the program, genetic analysis of extant ginseng populations, studies on their morphological features and mating system were carried out to understand the processes taking place in ginseng populations.
Ginseng is known to be a relic of the Tertiary era[2] and its prosperity in extant habitats depends on its individual ability to adapt to the environment. Estimation of genetic and morphological variability, heterogeneity of populations, and differentiation within and among populations are usually the best way to describe the state of an endangered species[5,6]. At the same time, genetic variation of plant populations is known to be largely determined by evolutionary history and mating system of the species[7]. Therefore, elucidation of these items would be useful for understanding both the current state and perspectives of natural ginseng populations. Moreover detailed study of the ginseng mating system and embryology is urgent for propagation of sampling ginseng plants in reintroduction centers. Significant items of the conservation program are an evaluation of the current state of ginseng populations, including numbers, age composition and viability, and analysis of spatial and temporal variability of coenopopulations. Nowadays ontogenetic analysis of populations is widely used for resource estimation of medicine plants[8,9]. Such investigations should be conducted to forecast the fate of the exhausted natural populations of Panax ginseng.
In this paper, we summarize the results of our investigations on the ginseng mating system, genetics and ontogenetic structure of its natural populations for evaluation of its current state and perspectives.
Material and methods
Sampled populations Wild-growing Panax ginseng plants were sampled from 3 non-protected natural populations: Khasan (Kh), Blue Mountain (BM) and Sikhote-Alin (SA). Collected living plants were transferred to an open experimental nursery in the natural ginseng habitats (Spassky District of the Primorsky Kray) for further investigations; a part of the plants was also transferred to the laboratory greenhouse.
Sample material Fresh leaves or those frozen in liquid nitrogen were used for allozyme, amplified fragment length polymorphism (AFLP), and simple sequence repeats (SSR) analyses. A total of 206 plants of wild Panax ginseng were studied with allozyme analysis (34 from Kh, 49 from BM and 123 from SA). For AFLP assay, 35 Panax ginseng plants were studied (10 from SA, 14 from BM, and 11 from Kh), and 57 plants were studied with SSR markers (20 from SA, 19 from BM, 18 from Kh).
For ontogenetic research, wild ginseng plants were pictured and their above-ground organs were collected, dried and studied.
Mating system experiments were conducted in the laboratory greenhouse and the nursery over 4 years. For pollination experiments and embryology studies, flowering 5–25-year-old ginseng plants from natural populations and 5-year-old cultivated plants were used.
Allozyme analysis Enzyme extraction, electrophoresis, enzyme staining and interpretation were carried out as described in Koren et al[10]. Enzymes were extracted from fresh leaves or those frozen in liquid nitrogen.
AFLP and SSR analysis Genomic DNA was extracted according to Echt et al[11] and then purified by Murray and Thompson[12].
Amplified fragment length polymorphism analysis was conducted according to Vos et al[13]. DNA was digested using the restriction endonucleases EcoRI and MseI. AFLP primers for pre-amplification contained 2 selective nucleotides. The EcoRI primers contained 3 selective nucleotids and 4 selective nucleotids were used for the MseI primers. AFLP adapters and primers were synthesized by “SibEnzim” (Russia). Selectively amplified products were analyzed using the ABI 3100 automated DNA sequencing system (Applied Biosystems, USA). Each of the EcoRI-NNN primer types was labeled with FAM fluorescent dye at the 3' end. Size alignment of the AFLP fragments was carried out with ABI GeneScan Analysis Software (Applied Biosystems) with the ABI GeneScan 500 LIZ internal lane size standard. DNA fragments ranging from 50 to 500 bp in size from the AFLP analysis were scored.
Primers designed to flank 2 microsatellite loci (CT12, CA33) isolated from the ginseng genome were 5'-GAGAG TAACC ACAGG ATAGA GAAA-3' (CT12 F), 5'-CTCCC TTTAC AGGTA GATAG TGAA-3' (CT12 R), and 5'-CGATG TGGAT TTCAA TTTTA AG-3' (CA33 F), 5'-GGTCT ATGAG CCTAG TTTTC ATG-3' (CA33 R)[14]. Reaction mixtures and PCR amplification profiles were carried out according to Qin et al[14].
Polymorphism was detected by automated capillary electrophoresis of fluorochrome-labeled polymerase chain reaction (PCR) products. The forward primers were synthesized and labeled with fluorochrome R110 or R6G by “Syntol” (Russia). Duplex PCR products were separated with an ABI 310 DNA sequencer (Applied Biosystems) and the fragments were sized by a ladder labeled with a fluorochrome LIZ. Bands used in scoring ranged in size from 50 to 500 bp.
Data analysis Presence/absence matrices (“1” for present, “0” for absent) for AFLP and SSR markers were generated for all scorable bands for each DNA sample. These matrices were analyzed by POPGENE[15] to estimate polymorphism parameters at both the population and species levels. An exact test for population differentiation[16] and Nei’s genetic distance and identity[17,18] were calculated for all pairwise combinations of populations by TFPGA[19]. Parameters of genetic diversity were estimated with the TFPGA[19] program from all loci studied including monomorphic ones. Dendrograms were constructed using UPGMA with TFPGA[19].
Mating system experiments For self-pollination and cross-pollination treatments, flowers were emasculated in buds and pollinated by mature pollen. A single pollen donor per plant was used in the outcross treatments. For agamospermy examination, all flowers were emasculated in buds and isolated from insects, so that seeds could result from agamospermy only. Intact ginseng plants were observed as a control. Fruits of all tested plants were harvested after maturation in the last week of September.
Pollen morphology and embryology Pollen from newly emerged anthers was fixed and uniformly stained with acetocarmine[20] to evaluate pollen size and fertility. Ovules were fixed in a mixture of formaldehyde and acetic acid under vacuum. Fixed ovules were dehydrated, processed through paraffin wax, sectioned at 5–8 µm thickness, stained with haematoxyline/eosin or with haematoxyline/alcian blue and visualized using a Zeiss Axioplan microscope. To study early stages of megagametophyte development we used the clearing technique for angiosperm ovules[21]. Examination of autofluorescence in ovules was carried out with a LSM 510 META confocal laser scanning microscope (Carl Zeiss, Germany) equipped with argon laser. Confocal z-stack images and single plans were obtained after excitation at 488 nm and emission at 522 nm using a 505-nm LP filter. The intensity of the Ar laser was 6%.
Ontogenetic study According to the Rabotnov–Uranov scheme[22], 9 different life stages were classified in the life history of Panax ginseng[23]: latent period: seeds (drupes); pregenerative (virgin) period: seedlings, juveniles, immatures, virgin young, virgin adults; generative period: generative young, generative medium, generative mature. Types of natural populations were determined according to Rysin and Kazantseva[24]. Terms for morphology description of plants were used following Serebryakov[25] and ontogenetic terminology[8].
Results
Genetic structure of extant ginseng populations
Electrophoretic analysis of the 25 Panax ginseng enzymes revealed gene products from 39 putative loci, only 3 of which were polymorphic. In general, the level of allozyme polymorphism was low for all ginseng populations studied. The lowest level of diversity was observed in SA (Table 1). The F-statistics analysis of the natural ginseng population (Table 1) indicates a decrease of total gene diversity in SA. FIT (individual inbreeding coefficient for the species) and FIS (individual inbreeding coefficient in respect to a sub-population) values demonstrate a small deficit of heterozygotes in all 3 populations studied, and the highest individual inbreeding coefficient for the species, FIT, is observed in Kh, indicating that processes of inbreeding and gene shift are more intense in this population than in 2 others. Proportion of the total diversity among sub-populations (FST), demonstrates a low level of subdivided ginseng populations, supposing the most part of a total genetic diversity is observed within sub-populations. The most level of subdividing is observed in SA, whereas there is no subdividing in BM.
Full table
All 4 labeled selective primers used for AFLP analysis revealed polymorphism (Table 2). Using DNA from 35 plants, the primers initiated the synthesis of 214 amplicons, of which 202 were polymorphic. It corresponds to a level of polymorphism of 94.39% (Table 3). Of the polymorphic bands, 112 were found in a sample of the SA, 167 in the BM and 161 in the Kh (Table 2). The indices of genetic variability in the SA were the lowest (Table 3).
Full table
Full table
Both microsatellite loci used in the study were polymorphic when considered for 57 representatives of 3 populations. A total of 14 alleles were detected over SSR primer sets; 7 for CT12 and 7 for CA33 (Table 2). The genetic diversity estimated with SSR markers was high and coincident with that obtained with AFLP markers (Table 3).
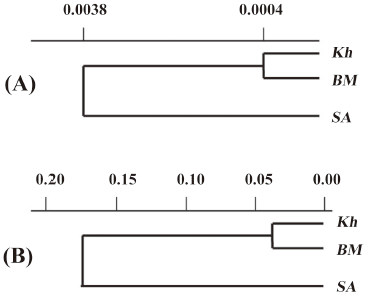
Using Nei’s genetic distance matrix based on allozyme and SSR data UPGMA dendrograms are constructed. According to both dendrograms, genetic distance between Kh and BM is very small, whereas distance between these 2 populations and SA is much greater (Figure 1).
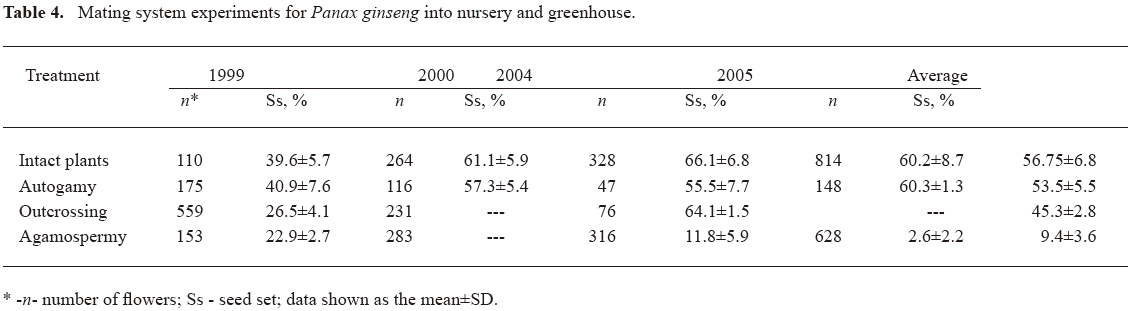
Mating system and developmental processes taking place in Panax ginseng Results of the mating system experiments are summarized in Table 4.
Full table
Ginseng pollen is prolate spheroidal in equatorial view and triangular in polar view, with slightly flattened poles. The mean pollen diameter was 27.4029±3.8151 µm. This value is close to the one published for Korean ginseng[26]. Division of the nuclei into pollen grains occurs before the anther undergoes dehiscence. Pollen grains fertility averaged 56%, highlighting disturbances in microsporogenesis.
Most Panax ginseng ovaries exhibited tetrasporic female gametophyte development. The ovary is 2-locular and each loculus contains 2 ovules. Upper loculus has a reduced embryo sac, but normal development of the upper ovule is rarely observed (Figure 2a). Normal embryo sac develops in the lower loculus. Often ginseng plants have formed fruits with one loculus. The differentiation of the archesporium in Panax ginseng occurs early.
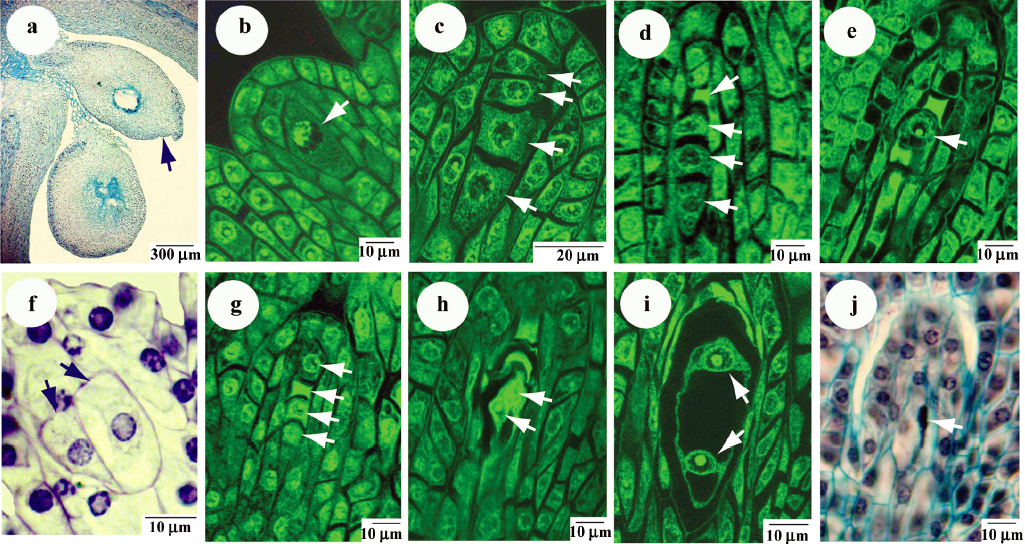
The polygonum-type of embryo sacs is developed in Panax ginseng’s ovules. The lower anatropous, tenuinucellate ovules bears a large central megaspore mother cell (MMC, Figure 2b). Meiotic division of MMC leads to 4 (or sometimes 3) megaspores arranged in a line (Figure 2c). After that, 3 (or 2) megaspores degenerate (Figure 2d) whereas the last one enlarges and becomes a functional megaspore with vacuolated cytoplasm (Figure 2e). Mature female gametophytes usually consist of 7 cells: 3 chalazal antipodals, a single large one with fused central nucleus near the egg apparatus, 2 synergids, and an egg cell at the mycropylar end.
In Panax ginseng, several deviations take place during megagametophyte development. Absence of the mother megaspore cell or its duplication are often observed (Figure 2f). Disturbances in succession of megaspores degeneration are revealed in Figure 2g. Sometimes there is no visible debris of degenerated megaspores during embryo sac formation. Nuclei are not separated at the beginning stages of embryo sac formation (Figure 2h, 2i). Very often the degeneration of all megaspores in ovule is occurred (Figure 2j) and embryo sac does not form. Megagametophytes development in different ovules of one ovary is asynchronous.
Double fertilization is asynchronous. After fertilization, the synergids degenerate and their debris is observed as 2 darkly stained bodies near the micropyle. The egg cell fertilization precedes the central cell fertilization by premitotic syngamy. The fusion of the first sperm cell with nucleus of the central cell of the embryo sac gives rise to the nuclear endosperm, followed by fertilization of the egg cell by the second sperm cell. The times of pollen tube outgrowth, fertilization and the resting period of the zygote are slightly different to the same data for Chinese ginseng[27]. Most of the normal developing ovules are fertilized at the post pollination stage for about 3 days. Rates of zygotic and parthenogenetic embryo development are different. After cross-pollination, embryos undergo a short period of dormancy.
The unfertilized egg cell and polar nucleus are not degenerated in the 10 days after flower opening.
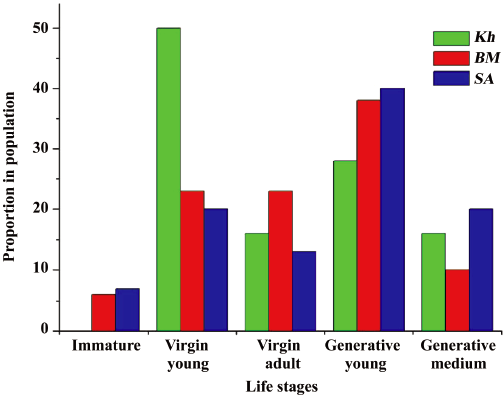
Ontogenetic structure of natural ginseng populations Life stage structure (ie, relative proportion of each life stage in the studied populations) is shown in Figure 3. These data were used to establish the types of populations and to evaluate their vital force as reflected in the Discussion.
Discussion
The evolutionary history of Panax ginseng is known to influence its genetic diversity[28]. Our data on allozyme polymorphism indicate that Russian’s ginseng populations have apparently experienced a severe genetic bottleneck during their evolutionary history. Although Panax ginseng is restricted to the ancient area where glaciation was absent, there have been several climatic fluctuations that would have affected the distribution of this species. In particular, during the last period of climate cooling (about 18 000 to 20 000 years ago) the territory of Central Sikhote-Alin was occupied with tundra phytocoenosis,[29] which was unfavorable for ginseng. Therefore, the lowest level of variation observed in SA could be attributed to the founder effect due to the lack of a Sikhote-Alin ginseng refugium during the recent period of climate cooling. In early Holocene (10 500 to 8000 years ago), intensive warming of the climate promoted the disappearance of permafrost in mountains and the expansion of thermophilic plants from southern refuges to the north[30,31]. Probably, recolonization of Sikhote-Alin by ginseng may have occurred in that period. This assumption is in full agreement with the computation of evolutionary time from the mean Nei’s genetic distance between populations on the base of allozyme data. According to Nei’s “molecular clock” supposed for allozyme data[32], DN=0.0016 obtained in our study corresponds to approximately 8000 years (considering the mean rate of mutation equal to 10-6).
The results of F-statistics analysis[33] show that genetic diversity is decreased from south-west, Kh, towards the north-east, for example SA (Table 1). Perhaps, the center of genetic richness and, possibly, the ginseng origin, was situated south-west of the location of populations studied in the present work. Therefore, study on the genetic diversity of ginseng plants growing outside Russia would be decisive to clear this presumption and improve our understanding of the evolutionary history of Panax ginseng.
The low level of allozyme polymorphism of natural ginseng populations is in agreement with results obtained recently by other techniques used for evaluating genetic polymorphism of Panax ginseng[1,34–36]. Allozyme analysis examines only the coding portion of the species DNA and therefore cannot cover the variability of the entire genome. In contrast, PCR-based techniques allow extensive genetic sampling over the entire genome and reveal as a rule the higher genetic variability. Automated capillary electrophoresis of fluorochrome-labeled PCR products is considered as the detecting system with the highest resolving power. The AFLP method[13] has the advantages of PCR technology and the stringency of the restriction fragment length polymorphism (RFLP) technique and therefore AFLP markers are highly reliable and reproducible ones. SSR markers were used in this research because they provide one of the most hypervariable and reproducible marker systems[37].
Exact test of differentiation of ginseng sample studied with SSR markers showed a highly significant difference between SA and BM and SA and Kh (P=0.0000), whereas differences in allele frequencies between the Kh and BM were not significant (P=1.0000). Exact test of population differentiation studied with AFLP markers showed a significant difference between the Kh and BM (P=0.0077), whereas differences in allele frequencies between the SA and BM and between SA and Kh were not significant (P=1.0000 and P=0.4531, respectively). Such genetic patterns demonstrate the discriminative power of SSR markers that exceed this ability of AFLP markers and testify to the heterogenity of the studied Panax ginseng sub-populations. The AFLP data allows us to classify the SA as the youngest among 3 populations under study; this is in full concordance with allozyme data.
Dendrograms constructed using allozyme and SSR markers were similar and showed that Kh and BM clustered together but SA joined them at a certain genetic distance, confirming the genetic remoteness of SA from Kh and BM (Figure 1). For an allozyme-based dendrogram, the mean Nei’s genetic distance (DN) between populations was 0.0016, whereas for the dendrogram based on SSR markers, the mean DN between populations was higher and amounted to 0.1258 (Figure 1). With AFLP markers the genetic remoteness of Kh plants was shown. It was reported that the type of molecular markers used influenced the patterns of relationships revealed in diversity studies[38,39]. Thus, dendrograms of genetic similarity of sorghum germplasm based on PstI/MseI AFLP markers produced clusters that were different to those based on EcoRI/MseI AFLP or SSR markers[39]. The marker sets that adequately cover the entire genome will provide a more precise estimate of genetic relationships[39].
According to AFLP, the index of population differentiation (Gst) was equal to 0.1484 (Table 3), which indicates on approximately 15% of genetic diversity among the ginseng populations. This is closely related to the pattern revealed by the allozyme analysis of wild Panax ginseng (Fst=0.1896)[40] and wild Panax quinquefolius (Gst=0.176)[41], and to the pattern obtained by RAPD for cultivated Panax quinquefolius (Gst=18%)[42]. According to SSR, the proportion of variation found among populations was equal to 0.2608 (Table 3) and was coincident with the population differentiation obtained with RAPDs for P. ginseng natural populations, Gst=23.9%[43], and for Panax quinquefolius natural populations, Gst=28%[43]. By the fluorescently-based automated AFLP method over 40% of the genetic variation was shown to be among populations of wild P. stipuleanatus[44].
Allozymes and RAPD assays have provided an average Gst<19% for outbreeding species, 21.2%-24.0% for species with a mixed mating system, and 58.7%-59.6% for inbreeding dicots[45,46]. At the same time, regardless of the breeding system, the total gene diversity at the species level was partitioned primarily between populations (Gst=0.556-0.924), as was shown by RAPD markers for 3 orchid species with different mating systems[47]. Therefore, a species breeding system often plays a central role in determining the distribution of genetic variation within and between populations[45], and other life history traits affect the species population structure too[47]. According to RAPD data, the value of Gst at the species level for Panax ginseng was 23.9%[43], indicating Panax ginseng possesses a mixed mating system.
The capability of ginseng to produce seeds via autogamy, outcrossing or agamospermy without pollination was shown early[48,49]. Because renewal of ginseng population is only possible via seed reproduction, this item required more detail research.
Seed set of autogamy experiments were comparable with controls over 4 years, demonstrating the absence of any self-incompatibility barrier in Panax ginseng. So, the capability of ginseng plants to cross- and self-pollinate is unlimited, although seed sets in the nursery are larger than in the greenhouse. Results of agamospermy tests confirm our assumption that Panax ginseng is a facultative apomictic plant with a diplospory type of agamospermy. This type of apomixis occurs without a reduction in chromosome number due to disturbances in meiosis and formation of diploid megaspores. In this work, we studied ginseng embryology to observe embryo development in different ways of seed formation.
Seed development in angiosperms is provided with 3 steps: the fertilized egg cell develops into the embryo, the fertilized central cell gives rise to the endosperm, and the ovule’s integument(s) form the seed coat. During sexual reproduction, these processes are dependent upon double fertilization, but for agamospermy, one or more of these developmental pathways are activated in the absence of fertilization and some anomalies in reproductive structures and embryogenesis are often observed in this case[50,51].
Agamospermy is common among the Araliaceae family; for example, it was shown experimentally in Acanthopanax sessiliflorum, Aralia mandshurica, Eleutherococcus senticosus[20,52]. A shift to higher levels of inbreeding or asexual reproduction is supposed to be often associated with a processes of rapid colonization and may be caused by selection[53,54]. Agamospermy in P. ginseng may be its ancient adaptation that does not play a significant role in seed reproduction nowadays. On the other hand, asexual reproduction may be a relatively recent acquirement of the species due to the low plant density within natural populations.
Gametophytic anomalies such as asynchronization and anomaly in embryological processes in one inflorescence, discords in developmental stages of embryo and endosperm in different ovules indicate the conversion of species into another avenue of seed reproduction[20,55]. These disturbances are considered to increase the evolution potential of species[56].
The reproductive system of ginseng possesses high plasticity and stability of fertilization processes that help it to survive in stress conditions (delay of pollination, unfavorable temperature). Disturbances caused by external or internal factors are reduced due to morphogenetic plasticity of ginseng plants and their capacity to vary the method of embryo development in agreement with these characteristics.
According to the coenopopulation analysis scheme, BM and SA are characterized as populations of perennials with moderate environmental conditions for seed reproduction (N3[23]), where the number of virgin plants is approximately equal to generative young and medium individuals. In life history structures of these populations, there are no individuals in the youngest life stages (seedlings and juveniles) and immature and generative mature individuals are rare. The generative young stage is predominant in BM and SA, whereas the virgin young stage prevails in Kh. So, Kh can be considered as a population being in unfavorable conditions for seed reproduction (N4[23]). As a whole, the analysis of life stage structure of ginseng populations revealed that virgin and generative (except generative mature stage) individuals were the most abundant in all habitats studied.
Analysis of life stages structure of studied ginseng populations demonstrates that they can be considered normal, but all of them are not full-constituent because some life stages are absent or occur rarely. This may be connected with ginseng roots being used as a raw material for medicine, when the whole plants that are mainly of the generative mature stage are extracted from populations continuously. Just this stage is lost in all populations studied. Moreover, renewal of ginseng populations happens via seed reproduction only. The pregenerative (virgin) period of Panax ginseng, (ie, the increase of vegetative weight) is very long; therefore, restoration of the life stage structure of the extant natural ginseng population requires much time. However, because all 3 populations studied are normal (viable), we hope that reintroduction of natural ginseng population is possible yet.
Genetic and population data presented in this paper characterize Panax ginseng rather controversially. On one hand, this plant demonstrates a high level of mating system plasticity, multiplicity of ways of reproduction, and an unprecedentedly wide interval of longevity. Such signs are usually inherent in flourishing species. However, this plant is very rare in natural habitats; it possesses only moderate genetic variability and its population structures are not full-constituent for all 3 populations studied. These last signs are indicators of depression. One can suppose that not only overharvesting but some organic defects in the plant body constitution or in its life strategy are responsible for such a complicated situation. The history of the Araliaceae family dates back to ancient times and some later biochemical inventions are absent in body of the ginseng plant. For example, cell wall structure is not sufficiently hard to stop the distribution of fungi or bacterium infection. Such deficiencies can be made up with more effective reproduction only partly since mechanisms of seed spread are restricted also. Some other antagonisms can be laid bare in further analysis. Therefore, only reintroduction oriented to overcome the diversity of shortcomings in ginseng genetics, biochemistry and life style can be successful. To fit these purposes, the stock material for reintroduction has to have the maximal genetic diversity possible for local population. It may be most important for the current state, the overharvesting and habitat-destroying human activity must be stopped and regulated [57,58].
Acknowledgements
We thank Drs VL SEMERIKOV, A BONDAR, and E BRENNER for their kind help in AFLP analysis.
References
- Zhuravlev YuN, Kozyrenko MM, Artyukova EV, Reunova GD, Muzarok TI, Elyakov GB. Genetic typing of Panax ginseng by use RAPD-PCR. Doklady RAS 1996;349:111-4. [In Russian].
- Grushwitsky IV. Ginseng: the aspects of biology. Leningrad: Nauka; 1961. [In Russian]
- Red Book of Russian Federation. Moscow: Rosagropromizdat; 1988. [In Russian]
- Zhuravlev YuN, Burundukova OL, Koren OG, Zaytseva YuA (Khrolenko YuA), Kovaleva LE. C.A. Meyer: Biodiversity evaluation and conservation. In: Bailey WG, Whitehead C, Proctor JTA, Kyle JT, editors. The challenges of the 21-st century. Proceedings of International Ginseng Conference. 1994. Vancouver, Canada. p 162–8.
- Brown AHD. Enzyme polymorphism in plant populations. Theor Pop Biol 1979;15:1-42.
- Soltis PS, Soltis DE. Genetic variation in endemic and wide-spread plant species: examples from Saxifragaceae and Polystichum (Dryopteridaceae). Aliso 1991;13:215-23.
- Hamrick JL, Godt MJV. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland: Sinauer Associates; 1989. p 43–64.
- Ontogenetic Atlas of Medicinal Plants. Ioshkar-Ola: Izd-vo Mar; 1997. p240. [In Russian]
- Vedernikova OP. Population-ontogenetic approach in estimation biology recourses of medicinal plants in republic Maryi Al. In: Botanical Studies in the Asian part of Russia. v 3. Barnaul. 2003. p 9–10. [In Russian]
- Koren OG, Potenko VV, Zhuravlev YuN. Inheritance and variation of allozymes in Panax ginseng C.A. Meyer (Araliaceae). Int J Plant Sci 2003;164:189-95.
- Echt CS, Erdahl LA, McCoy TJ. Genetic segregation of random amplified polymorphic DNA in diploid cultivated alfalfa. Genome 1992;35:84-7.
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 1980;8:4321-5.
- Vos PR, Hogers M., Bleeker M, Reijans M, van der Lee T, Homes M, et al. AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 1995;23:4407-14.
- Qin J, Leung FC, Fung Y, Zhu D, Lin B. Rapid authentication of ginseng species using microchip electrophoresis with laser-induced fluorescence detection. Anal Bioanal Chem 2005;381:812-19.
- Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian J Botany 1997;129:157.
- Raymond ML, Rousset F. An exact test for population differentiation. Evolution 1995;49:1280-3.
- Nei M. Genetic distance between populations. Amer Naturalist 1972;106:283-92.
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978;89:583-90.
- Miller MP. Tools for population genetic analysis (TFPGA) 1.3: A Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author; 1997.
- Hohlov SS. The revelation of apomixis in the flora of flowering plans of USSR. Saratov: Nauka; 1978. [In Russian]
- Stelly DM, Peloquin SJ, Palmer RG, Crane CF. Mayer’s hemalum-methyl salicylate: A stain-clearing technique for observation within whole ovules. Stain Technology 1984;59:155-61.
- Rabotnov TA. On coenopopulations of perennial herbaceous plants in natural coenosis. Vegetatio 1969;19:87-95.
- Khrolenko YuA, Burundukova OL, Bezdeleva TA, Muzarok TI, Zhuravlev YuN. Age stages in the ontogeny of cultivated Panax ginseng C.A. Mey. Biol Bull 2007;2:120-5.
- Rysin LP, Kazantseva TN. Method of coenopopulation analysis in geo-botanical research. Bot Zhurnal 1975;60:199-209. [In Russian].
- Serebryakov IG. Morphology of vegetative organs in higher plants. M.: Sovetskaya nauka; 1952. [In Russian]
- Wen J, Nowicke JW. Pollen ultrastructure of Panax (the ginseng genus, Araliaceae), an eastern Asian and eastern North American disjunct genus. Am J Bot 1999;86:1624.
- Yong-quan L, Jia-heng S. Embryological studies on ginseng (Panax ginseng C.A. Mey). Acta Bot Sin 1989;31:653-60.
- Hamrick JL, Godt MJV, Sherman-Broyles SL. Factors influencing levels of genetic diversity in woody plant species. New Forests 1992;6:95-124.
- Golubeva IV, Karaulova LP. Vegetation and climatostratigraphy of pleistocene and holocene of the USSR Far East South. Moscow: Nauka; 1983. [In Russian]
- Korotkii AM, Lobanova LA. On the rate and conditions of Holocene peat accumulation in the Far East. In: Paleography analysis and stratigraphy of antropogen of Far East. Vladivostok; 1983. p 109–19. [In Russian]
- Korotkii AM, Grebennikova TA, Pushkar VS, Razzhigaeva NG, Volkov VG, Ganzei LA, Climatic changes on south of the Far East in the late neozoic period (Miocene-Pleistocene). Vladivostok: Far East State University Press; 1996. [In Russian]
- Nei M. Interspecific gene differences and evolutionary time estimated from electrophoretic data on protein identity. Amer Naturalist 1971;105:385-98.
- Wright S. The genetic structure of population. Ann Eugen 1951;15:323-54.
- Wen J, Zimmer GA. Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol Phylogenet Evol 1996;6:167-77.
- Zhao SJ, Zhao YH, Yang ZT. Genetic analysis of ginseng germplasm by lactate polyacrylamide gel electrophoresis of seed protein. J Ginseng Res 1998;22:168-72.
- Kim Ch, Choi H-K. Genetic diversity and relationship in Korean Ginseng (Panax schinseng) based on RAPD analysis. Korean J Genet 2003;25:181-8.
- Fossati T, Grassi F, Sala S, Castiglione S. Molecular analysis of natural populations of Populus nigra L. intermingled with cultivated hybrids. Mol Ecol 2003;12:2033-43.
- Parsons BJ, Newbury HJ, Jackson MT, Ford-Lloyd BV. Contrasting genetic diversity relationships are revealed in rice (Oryza sativa L.) using different marker types. Mol Breed 1997;3:115-25.
- Menz MA, Klein RR, Unruh NC, Rooney WL, Klein PE, Mullert JE. Genetic diversity of public inbreds of sorgum determined by mapped AFLP and SSR markers. Crop Sci 2004;44:1236-44.
- Zhuravlev YuN, Koren OG, Reunova GD, Artyukova EV, Kozyrenko M. Ginseng conservation program in Russian Primorye: genetic structure of wild and cultivated populations. J Ginseng Res 2004;28:60-6.
- Cruse-Sanders JM, Hamrick JL. Genetic diversity in harvested and protected populations of wild American ginseng, Panax quinquefolius L. (Araliaceae). Am J Bot 2004;91:540-8.
- Schluter C, Punja ZK. Genetic diversity among natural and cultivated field populations and seed lots of American ginseng (Panax quinquefolius L.) in Canada. Int J Plant Sci 2002;163:427-39.
- Zhuravlev YuN, Reunova GD, Kats IL, Muzarok TI. Analysis of genetic variability in populations of C.A. Meyer using RAPD, ISSR and AFLP markers. In: Oh S, Choi KT, editors. Advances in Ginseng Research 2006. Proceedings of the 9th International Symposium on Ginseng; 2006 Sep 25–28; Geumsan, Korea. p 423–40.
- Zhou SL, Xiong GM, Li ZY, Wen J. Loss of genetic diversity of domesticated Panax notoginseng F H Cheng as evidenced by ITS sequence and AFLP polymorphism: a comparative study with P. stipuleanatus H T et K M Feng. J Integr Plant Biol 2005;47:107-15.
- Hamrick JL, Godt MJW. Conservation genetics of endemic plant species. In: Avise JS, Hamrick JL, editors. Conservation genetics, case histories from nature. New York: Chapman and Hall; 1996. p 281–304.
- Bussell JD. The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petraea (Lobeliacese). Mol Ecol 1999;8:775-89.
- Sun M, Wong KC. Genetic structure of three orchid species with contrasting breeding system using RAPD and allozyme markers. Amer J Bot 2001;88:2180-8.
- Koren OG, Krylach TYu, Zaytseva YuA, Zhuravlev YuN. 1998. Floral biology and embryology of C.A. Meyer. In: Weber HCh, Zeuske D, Imhof S, editors. Ginseng in Europe. Proceedings of the 1st European Ginseng Congress; 1998 Dec 6–11; Marburg, Germany. Marburg: Philipps-Univ; 1998. p 221–31.
- Koren OG, Gorpenchenko TYu, Muzarok TI, Zhuravlev YuN. Genetic approaches for the problem of monitoring and management of forest ginseng ( C.A. Meyer): using allozyme markers. Proceedings of the International Symposium for the Sustainable Forest Ginseng Production. 2007 Nov 9–10; Sangju, Republic of Korea. Sangju: Sangju National University; 2007. p 83–97.
- Petrov DF. Genetically regulating apomixis. Novosibirsk: Nauka; 1979. [In Russian]
- Koltunow AM. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 1993;5:1425-37.
- Petrov DF. Progeny without fathers (apomixis and its significance for breeding). Novosibirsk: Nauka; 1976. [In Russian]
- Barrett SCH, Husband B. The genetics of plant migration and colonization. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland: Sinauer Associates; 1990. p 254–77.
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity 1997;78:311-27.
- Nogler GA. Gametophytic apomixes. In: Johry BM, editor. Embryology of Angiosperms. Berlin; 1984. p 475–510
- Shishkinskaya NA, Udakova OI, Tyrnov VS. Population embryology and apomixis of cereals. Saratov: Saratov University Press; 2004. [In Russian]
- Zhuravlev YuN, Koren OG, Kozyrenko MM, Reunova GD, Artyukova EV, Krylach TYu, Use of molecular markers to design the reintroduction strategy for . In: Chou CH, Waller GR, Reinhardt C, editors. Biodiversity and allelopathy: from organisms to ecosystems in the Pacific. Taipei: Academia Sinica; 1999. p 183–92.
- Zhuravlev YuN, Koren OG, Reunova GD, Artyukova EV, Kozyrenko MM, Muzarok TI, et al. Ginseng conservation program in Russian Primorye: genetic structure of wild and cultivated populations. J Ginseng Res 2004;28:60-6.