Inhibition of acid-induced apoptosis by targeting ASIC1a mRNA with short hairpin RNA1
Introduction
Acid-sensing ionic channels (ASIC) are H+-gated cation channels expressed throughout mammalian central and peripheral nervous systems[1–6]. They belong to the degenerin/epithelial sodium channel superfamily. Until now, 6 ASIC subunits have been cloned: ASIC1a[1], ASIC1b[2], ASIC2a[3], ASIC2b[7], ASIC3[1,2,8], and ASIC4[9]. Functional ASIC are usually tetrameric assemblies of these ASIC subunits in homomeric or heteromeric conformation[10]. However, neither ASIC2b nor ASIC4 can form functional homomeric channels themselves[7,9,11]. ASIC2b has been shown to associate with other subunits and modulate their activity[7]. Various ASIC exhibit different kinetics, pH sensitivities, and tissue distribution[7,10,12]. The pH of half-maximal activation (pH0.5) of these channels differs: ASIC1a (or ASIC1), pH0.5=6.2[1]; ASIC1b, a splice variant of ASIC1a, pH0.5=5.9[13]; ASIC2a, pH0.5=4.4[10]; and ASIC3, pH0.5=6.5[2]. Besides Na+ permeability, homomeric ASIC1a can flux Ca2+[1,14,15].
Both ASIC1a and ASIC2a subunits have been shown to be abundant in the brain, whereas ASIC2b is present in both brain and sensory neurons[5,16–18]. ASIC4 is found to be co-expressed with other subunits in many areas of the brain. ASIC3 is expressed almost exclusively in sensory neurons in rats and generates a biphasic current with a fast, inactivating phase, followed by a sustained component[2,4,12].
Under most pathological conditions ranging from ischemia, inflammation, epilepsy to cancer, tissue pH often falls to about pH 6.0[19-21]. Acidosis is assumed to play a critical role in cell injury[21]. Although the detailed mechanisms remain unclear, ASIC are thought to be involved in this process. In sensory neurons, ASIC have been proven to play an important role in nociception during a tissue acidosis, for instance in muscle and cardiac ischemia[22] and in inflammation[23]. In peripheral sensory neurons, ASIC have been implicated in mechanosensation[24] and perception of pain during tissue acidosis[25-29], particularly in ischemic myocardium where ASIC are likely transducers of anginal pain[30]. The presence of ASIC in the brain, which lacks nociceptors, suggests that these channels have functions beyond nociception. For example, our previous studies have identified the characteristic of ASIC in the hippocampal neurons[31]. Now, increasing evidence has proved that ASIC1a is involved in synaptic plasticity, learning/memory, and fear conditioning[18,32] and participates in neuronal damage associated with tissue acidosis[15,33].
Now, ASIC have been found in malignant gliomas[34]. Recent studies have revealed that the surface expression of ASIC2 could inhibit the amiloride-sensitive current and migration of glioma cells[34]. However, the role of ASIC1a in acid-induced cytotoxicity remains undefined. Here, using the specific technology of RNAi (RNA interference), in vitro cell toxicity assays, and an apoptosis study, we found that extracellular acid induced cell death and apoptosis in the rat C6 glioma cell line. Data also showed that reducing ASIC1a proteins by stable expression of short hairpin RNA (shRNA) significantly decreased acid-induced cytotoxicity.
Materials and methods
Chemicals and antibodies Propidium iodide (PI) and 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) were purchased from Sigma Co (St Louis, MO, USA). The antibody against ASIC1a (category N
Cell culture The rat glioma C6 cell line derived from rat neurogliocytoma was purchased from the Cell Culture Centre of the Institute of Basic Medical Sciences (Chinese Academy of Medical Sciences, Beijing, China). The C6 cell line was cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, USA), 100 U/mL penicillin, 100 μg/mL streptomycin, 0.03% L-glutamine (w/v), and 2.2% sodium bicarbonate (w/v). The ASIC1a-silenced cells and the control cells were screened with the G418. The cells were cultured at 37 °C a in a humidified atmosphere with 5% CO2 (310/Thermo, Forma Scientific, Marietta, OH, USA). G418, penicillin, L-glutamine, nimodipine, (+/–)-5-methyl-10,11-dihydro-5H-dibenzo[a,d] cyclohepten-5,10-imine (MK801), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), and all other reagents were purchased from Sigma (St Louis, MO, USA).
Analysis of cell death The cells were plated in 96-well plates at a density of 1×104 cells/well and incubated for 16–20 h. Then the cells were treated with acid incubation for 3 h in RPMI-1640 medium with 10 μmol/L MK801, 20 μmol/L CNQX, and 5 μmol/L nimodipine, which were added to eliminate the effect of glutamate receptors and voltage-gated Ca2+ channels. After drug treatment, the cells were washed twice with phosphate buffered saline (PBS) and were recultured in complete RPMI-1640 medium containing 10% FBS for about 4 h. Subsequently, the medium was replaced and the cells were incubated with 0.5 mg/mL MTT in complete RPMI-1640 medium for 4 h[15]. The surviving cells converted MTT to formazan, which generates a blue–purple color when dissolved in DMSO. The intensity was measured at 490 nm using a plate reader (Molecular Dynamics Inc, Sunnyvale, CA, USA) for the enzyme-linked immunosorbent assays. The relative percentage of survival was calculated by dividing the absorbance of the treated cells by that of the control in each experiment.
Cell injury assay
Lactate dehydrogenase (LDH) release assay The degree of cell injury was assessed by a quantitative measurement of released lactate dehydrogenase (LDH). The cells were treated with acid incubation as described earlier. Then the culture medium was removed and the cells were incubated with necessary cofactors as described by the manufac-turer’s assay kit (Sigma Diagnostics, St Louis, MO, USA). One percent of Triton X-100 in PBS was used to extract the remaining LDH from the cultures. Absorbance measurements at 440 nm were taken every 30 s for 3 min by using a 96-well microplate reader (Molecular Dynamics, USA). The resulting slopes defined the LDH units per sample. Data were expressed as a ratio of LDH released/(LDH released+LDH Triton-X extracted).
Apoptosis assay After exposure to the extracellular solution (pH 6.0) for 3 h, the cells continue to be cultured in the extracellular solutions (pH 7.4) for about 4 h[15]. In order to eliminate the effect of glutamate receptors and voltage-gated Ca2+ channels, MK801 (10 μmol/L), CNQX (20 μmol/L), and nimodipine (5 μmol/L) were also added to these solutions. Then the cells were washed with cold PBS and resuspended with binding buffer [10 mmol/L hydroxyethyl piperazine ethanesulfonic acid (HEPES)/NaOH (pH 7.4), 140 mmol/L NaCl, and 2.5 mmol/L CaCl2]. The solution containing 105 cells was transferred to a 5 mL tube, and 5 μL of Annexin-V–fluorescein isothiocyanate (FITC) and 10 μL PI were added to the culture medium and incubated for 30 min in the dark. Binding buffer was then added to each tube and the apoptotic cells were assessed with the flow cytometer.
Cell cycle analysis The cells were cultured and treated as indicated earlier. Then the cells were harvested, washed with PBS, and fixed in 70% ethanol. After being fixed for 12 h at –20 °C, the cells were collected by centrifugation and resuspended in 1 mL staining solution containing 50 μg/mL PI, 1 mg/mL RNase A, and 1 mg/mL glucose in PBS. The cells were analyzed for DNA content using a XL flow cytometer (Beckman Coulter Inc, Miami, FL, USA) after being incubated for 1 h of staining.
Western blot analysis Western blot analyses of ASIC1a were performed using the antibody against ASIC1a (category N
RT–PCR Total RNA was purified with RNeasy columns (Qiagen, Valencia, CA, USA). One-step RT–PCR was performed in 50 μL reactions using 100 ng RNA with 0.5 μmol/L of each primer. The annealing temperature was set at 53 °C and repeated for 25 cycles. Other PCR conditions and reagents were supplied and recommended by the manufacturer’s protocol for the Titan one-step system (Roche Applied Science, Indianapolis, IN, USA). The primer sequences[2,4,5,8,9,11,12] for ASIC1a were as follows: 5'-C
shRNA design and preparation The design of small interference RNA (siRNA) was based on Dharmacon siDESIGN Center software (Dharmacon., Lafayette, CO, USA). The sequence for ASIC1a RNAi was 5'-GATCCCGCGTGAATTCTACGACAGATTCAAGAGATCTGTCGTAGAATTCACGCTTTTT-3' (forward) and 5'-AGCTAAAAAGCGTGAATTCTACGACAGATCTCTTGAATCTGTCGTAGAATTCACGCGG-3' (reverse) and the sequence for the control RNAi was 5'-GATCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTT-3' (forward) and 5'-AGCTAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAGG-3' (reverse). The shRNA were synthesized in desalted and purified form by Dharmacon Research (Lafayette, CO, USA).
Construction of plasmid vectors and transfection The synthesized shRNA fragment containing the open reading frame of interfering-ASIC1a expression cDNA was inserted into the HindIII site of the pGCsi–U6–Neo–GFP cloning vector. The insert was digested with BamH I and HindIII and subcloned into the BamHI–HindIII restriction sites of the pGCsi expression vector. The C6 cells were transfected with the expression plasmids using Lipofectamine 2000 reagent (Invitrogen, USA), according to the manufacturer’s instructions. After transfection, the cells were subjected to the generation of the stable shRNA-transfected C6 cell line.
Generation of stable shRNA-transfected C6 cell line The rat shRNA-transfected plasmid for silencing the ASIC1a gene was earlier described. After transfection, the individual G418-resistant clones were selected. Under the G418 (800 μg /mL) selection, the cells were screened for 20 generations and confirmed to build the stable ASIC1a-knockdown C6 cell line with Western-blotting. After selection with G418, a stably transfected cell line of the ASIC-knockdown C6 cell line was obtained.
Statistical analysis Data were presented as mean ± SEM. Values were compared using either Student’s t-test or ANOVA. Two-way ANOVA was used to separate the effects of 2 variables and determine their interaction. P<0.05 was considered statistically significant.
Results
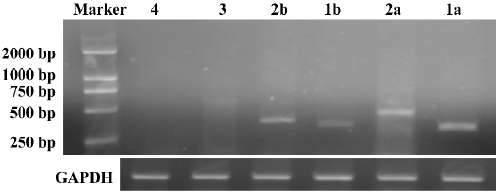
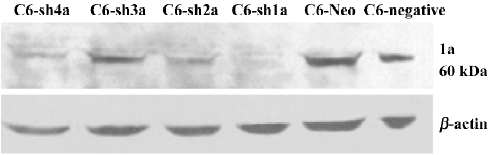
Successful build of the ASIC1a-silenced C6 cell line In the rat C6 glioma cell line, the transcription level of ASIC1a, ASIC1b, ASIC2a, and ASIC2b was identified with RT–PCR. The transcription of ASIC3 and ASIC4 was not detected in the cell line (Figure 1). To elucidate the role of ASIC1a in the acidosis, a U6 promoter-driven RNAi was used to silence the transcription of the ASIC1a gene in the C6 glioma cells. We designed and prepared the shRNA sequences directly against ASIC1a, and the control sequence was designed according to the Dharmacon siDESIGN Center software. Then we constructed the plasmid vectors with the pGCsi–U6–Neo–GFP cloning vector. The C6 cells were cotrans-fected with pGCsi–U6–Neo–GFP and either an ASIC1a-2specific RNAi or a control RNAi expression vector. Stably-transfected clones were selected with G418 and were screened for ASIC1a protein knockdown (C6–sh1-4a). Western blotting showed that a dramatic decrease in ASIC1a protein expression occurred in 1 clone from ASIC1a-specific RNAi (C6–sh1a) compared with those transfected with the vector only (C6–Neo) or the control RNAi clones (C6-negative) (Figure 2).
Knockdown of ASIC1a attenuated cytotoxicity induced by extracellular acid In the present study, we tested whether ASIC1a was involved in the neuroglioma cell toxicity when treated with different pH acidic solution. We applied acidic solutions with pH 6.0, pH 6.5, pH 6.9, and pH 7.4 to the C6–sh1a cells and the C6-negative cells, respectively. Cell survival was measured with the MTT assay, while cell injury was tested by LDH release. The experiments were conducted following the earlier-mentioned experiment procedure. To compare the relative percentage of survival conveniently, we took the absorbance of the C6–sh1a cells at pH7.4 as the control. The percentage of cell survival was calculated by dividing the absorbance of the treated cells by that of the control in each experiment. The extracellular acid solution induced cell death in both cell lines. As the pH value decreased, the percentage of survival for the two cell types decreased subsequently (two-way ANOVA, P<0.05), which demonstrated that acid stimulus played an important role in cytotoxicity. However, the percentage of survival had obvious distinction between different cell types (two-way ANOVA, P<0.05). It indicated that ASIC1a was involved in the cytotoxicity caused by the acid (Figure 3A).
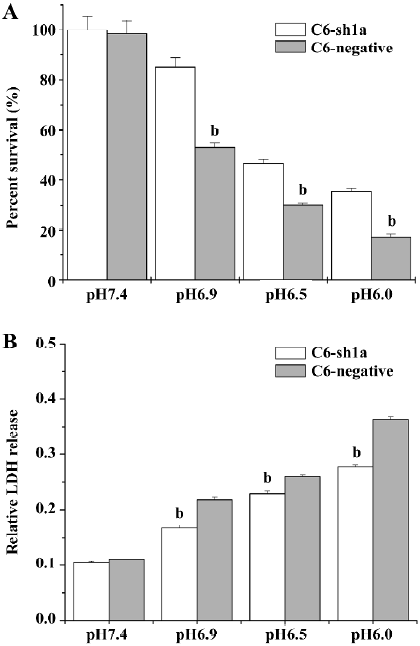
To assay the effect of pH on cell injury, the cells were treated as described earlier. After acid stimulus, the LDH release was measured 24 h later. It was found that exposure to acidic solutions (pH<7.4) induced LDH release in both types of cells compared with exposure to pH 7.4 solution (two-way ANOVA, P<0.05; Figure 3B). However, the LDH release in the C6–sh1a cells was much lower compared with that in the C6-negative cells (two-way ANOVA, P<0.05; Figure 3B) when treated with incubation in the same acid solution (pH<7.4). It also showed that the cytotoxicity caused by the acid was significantly attenuated by the knockdown of ASIC1a in the C6–sh1a cell line.
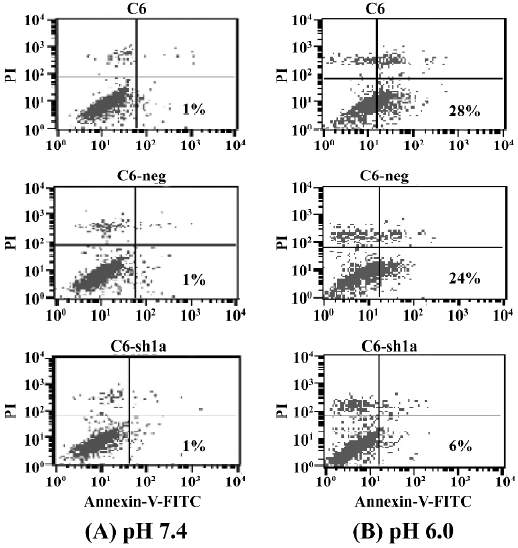
Knockdown of ASIC1a decreased the percentage of apoptosis induced by extracellular acid From the above experiments, we found that exposure to acidic solutions increased the cell death of the C6-negative and the C6–sh1a cells. As the percentage of survival of the C6–sh1a cells was much higher than that of the C6-negative cells, it indicated the ASIC1a was involved in the cytotoxicity induced by extracellular acid. We further assessed apoptosis from the cells that had been exposed to extracellular acidic solution. Experiments were conducted according to the earlier-mentioned experiment procedure. Apoptosis was assessed by Annexin-V–FITC staining to detect phosphatidylserine on the cell surface in conjunction with PI staining. Figure 4 shows the result of staining with Annexin-V–FITC and PI on the apoptotic cells and the normal cells (Figure 4). The percentages of apoptosis were very low with the normal pH 7.4 solution (about 1%) and increased with the pH 6.0 solution among the 3 cell types (Figure 4). Further, the C6–sh1a cells showed a decrease in Annexin V-positive and PI-negative cells (6%) when compared with the C6 cells and C6-negative control cells (28% and 24%, respectively). These results indicated the extracellular acid might induce apoptosis by activating ASIC1a.
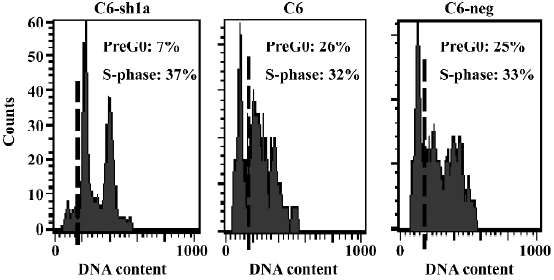
Cell death induced by acid was associated with the induction of cellular apoptosis To explore the mechanism of low cytotoxicity in ASIC1a-silenced cells exposed to extracellular acid, a cell cycle analysis was also performed. All 3 cell lines were treated with the same acid solution (pH 6.0) and experiments were conducted following the earlier-mentioned experiment procedure. As shown in Figure 5, the number of cells in the S-phase was 37%, 32%, and 33% for the C6–sh1a, C6, and C6-negative cells, respectively. In normal conditions (pH 7.4), all 3 cell lines had a normal cell cycle distribution with little subdiploid DNA (no more than 1%). Compared with the C6 and C6-negative cells, the C6–sh1a cells showed a remarkable decrease (from 26% to 7%) in the subdiploid DNA (sub-G0/G1) content, which suggested that knockdown of ASIC1a decreased cell apoptosis in the acid solution (pH 6.0) (Figure 5). Based on the results from the MTT and LDH experiments, it also indicated that the cell necrosis induced by acid was associated at least in part with the induction of cellular apoptosis.
Discussion
To date, 4 ASIC genes (ASIC1–4) have been identified, among which ASIC1a, ASIC2a, and ASIC2b all have a widespread distribution pattern in the brain. With the use of RT–PCR, we found that ASIC1a, ASIC2a, and ASIC2b were transcribed at high levels in the C6 cell line, which might be the result of their generation from neuroglioma. ASIC3 and ASIC4 were not detected, possibly because of their relatively low expression level in the C6 cell line.
In the present study, we studied the function of ASIC1a with the newly-developed RNAi technique. With the stably ASIC1a-silenced C6 cell line, we found that the extracellular acid could induce apoptosis via ASIC1a, which not only suggested that ASIC1a was involved in the apoptosis under the extracellular acid, but also indicated the main reason for cell death.
As we know, with the use of this double-staining regime, it is difficult to distinguish late apoptotic cells from necrotic cells in two-color flow cytometry. In fact, these cells are usually considered as dead cells with flow cytometry or MTT. In our study, the populations of cell death were tested 24 h after the cells were exposed to the sustaining extracellular acid stimulation for 3 or 4 h. However, when we analyzed the 3 populations of cells (non-apoptotic cells, early apoptotic cells, and necrotic cells or late apoptotic cells) only 6 h after exposure to an extracellular acid environment (pH 6.0), nearly one-third of the cells were apoptotic (32%). It was likely that many of the cells would become late apoptotic and dead cells. Thus, it suggested that silencing ASIC1a not only protected the cell from death, but also from apoptosis. It was also likely that ASIC1a was just like a dead receptor in the cellular membrane.
RNAi silences specific gene expression by targeting the relative mRNA and preventing it from being translated into a protein. Our studies were the first to utilize the highly-specific technology of RNAi to directly demonstrate that extracellular acid induced cytotoxicity and apoptosis by activating ASIC1a. As using ASIC1a-deficient mice has been limiting and difficult, the use of ASIC1a shRNA will greatly facilitate the precise identification of the ASIC1a gene function. RNAi mediated by shRNA is a powerful technology that allows the silencing of genes with great specificity and potency. Now it has been used more and more extensively in plants, nematodes, Drosophila, and mammalian cells to determine gene function. There are increasing reports on the use of siRNA in vitro[36–39] and in vivo[40–43]. Although more studies should be done to clarify whether ASIC1a-specific shRNA delivery in vivo is biologically effective, our data have shown that the inhibition of ASIC1a expression with RNAi was cytoprotective in vitro, which might have significant implications for future therapeutic interventions. As acid-induced cytotoxicity was critical to the pathogenesis of ischemia, inflammation, and tumors in the brain, it was suggested that shRNA for ASIC1a potentially have therapeutic value for which specific, safe antagonists do not currently exist.
References
- Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature 1997;386:173-7.
- Waldmann R, Bassilana F, de Weille J, Champigny G, Heurteaux C, Lazdunski M. Molecular cloning of a non-inactivating proton-gated Na channel specific for sensory neurons. J Biol Chem 1997; 272: 20 975–8.
- Waldmann R, Lazdunski M. H. +-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol 1998;8:18-24.
- Alvarez de la Rosa D, Zhang P, Shao D, White F, Canessa CM. Functional implications of the localization and activity of acid-sensitive channels in rat peripheral nervous system. Proc Natl Acad Sci USA 2002;99:2326-31.
- Alvarez de la Rosa D, Krueger SR, Kolar A, Shao D, Fitzsimonds RM, Canessa CM. Distribution, subcellular localization and ontogeny of ASIC1 in the mammalian central nervous system. J Physiol 2003;546:77-87.
- Krishtal O. The ASICs: signaling molecules? Modulators? Trends Neurosci 2003;26:477-83.
- Lingueglia E, de Weille JR, Bassilana F, Heurteaux C, Sakai H, Waldmann R, . A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem 1997; 272: 29 778–83.
- Babinski K, Le KT, Seguela P. Molecular cloning and regional distribution of a human proton receptor subunit with biphasic functional properties. J Neurochem 1999;72:51-7.
- Akopian AN, Chen CC, Ding Y, Cesare P, Wood JN. A new member of the acid-sensing ion channel family. Neuroreport 2000;11:2217-22.
- Waldmann R, Champigny G, Lingueglia E, De Weille JR, Heurteaux C, Lazdunski M. H. +-gated cation channels. Ann N Y Acad Sci 1999;868:67-76.
- Grunder S, Geissler HS, Bassler EL, Ruppersberg JP. A new member of acid-sensing ion channels from pituitary gland. Neuroreport 2000;11:1607-11.
- Babinski K, Catarsi S, Biagini G, Seguela P. Mammalian ASIC2a and ASIC3 subunits co-assemble into heteromeric proton-gated channels sensitive to Gd. J Biol Chem 2000; 275: 28 519–25.
- Chen CC, England S, Akopian AN, Wood JN. A sensory neuron-specific, proton-gated channel. Proc Natl Acad Sci USA 1998; 95: 10 240–5.
- Chu XP, Miesch J, Johnson M, Root L, Zhu XM, Chen D, et al. Proton-gated channels in PC12 cells. J Neurophysiol 2002;87:2555-61.
- Yermolaieva O, Leonard AS, Schnizler MK, Abboud FM, Welsh MJ. Extracellular acidosis increases neuronal cell calcium by activating acid-sensing ion channel 1a. Proc Natl Acad Sci USA 2004;101:6752-7.
- Price MP, Snyder PM, Welsh MJ. Cloning and expression of a novel human brain Na+ channel. J Biol Chem 1996;271:7879-82.
- Bassilana F, Champigny G, Waldmann R, de Weille JR, Heurteaux C, Lazdunski M. The acid-sensitive ionic channel subunit ASIC and the mammalian degenerin MDEG form a heteromultimeric H-gated Na channel with novel properties. J Biol Chem 1997; 272: 28 819–22.
- Wemmie JA, Chen J, Askwith CC, Hruska-Hageman AM, Price MP, Nolan BC, et al. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron 2002;34:463-77.
- Olson TH, Riedl MS, Vulchanova L, Ortiz-Gonzalez XR, Elde R. An acid sensing ion channel (ASIC) localizes to small primary afferent neurons in rats. Neuroreport 1998;9:1109-13.
- Nedergaard M, Kraig RP, Tanabe J, Pulsinelli WA. Ethylisopro-pylamiloride-sensitive pH control mechanisms modulate vascular smooth muscle cell growth. Am J Physiol Cell Physiol 1991;260:581-8.
- Siesjo BK, Katsura K, Kristian T. Acidosis-related damage. Adv Neurol 1996;71:209-33.
- Pan HL, Longhurst JC, Eisenach JC, Chen SR. Role of protons in activation of cardiac sympathetic C-fibre afferents during ischaemia in cats. J Physiol (Lond) 1999;518:857-66.
- Reeh PW, Steen KH. Tissue acidosis in nociception and pain Prog Brain Res 1996;113:143-51.
- Price MP, Lewin GR, McIlwrath SL, Cheng C, Xie J, Heppenstall PA, et al. The mammalian sodium channel BNC1 is required for normal touch sensation. Nature 2000;407:1007-11.
- Bevan S, Yeats J. Protons activate a cation conductance in a sub-population of rat dorsal root ganglion neurons. J Physiol 1991;433:145-61.
- Krishtal OA, Pidoplichko VI. A receptor for protons in the membrane of sensory neurons may participate in nociception. Neuroscience 1981;6:2599-601.
- Ugawa S, Ueda T, Ishida Y, Nishigaki M, Shibata Y, Shimada S. Amiloride-blockable acid-sensing ion channels are leading acid sensors expressed in human nociceptors. J Clin Invest 2002;110:1185-90.
- Sluka KA, Price MP, Breese NM, Stucky CL, Wemmie JA, Welsh MJ. Chronic hyperalgesia induced by repeated acid injections in muscle is abolished by the loss of ASIC3 but not ASIC1. Pain 2003;106:229-39.
- Chen CC, Zimmer A, Sun WH, Hall J, Brownstein MJ, Zimmer A. A role for ASIC3 in the modulation of high-intensity pain stimuli. Proc Natl Acad Sci USA 2002;99:8992-7.
- Benson CJ, Eckert SP, McCleskey EW. A possible mediator of myocardial ischemic sensation. Circ Res 1999;84:921-8.
- Weng XC, Zheng JQ, Gai XD, Li J, Xiao WB. Two types of acid sensing ion channels expressed in rat hippocampal neurons. Neurosci Res 2004;50:493-9.
- Wemmie JA, Askwith CC, Lamani E, Cassell MD, Freeman JHJ, Welsh MJ. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear condi-tioning. J Neurosci 2003;23:5496-502.
- Xiong ZG, Zhu XM, Chu XP, Minami M, Hey J, Wei WL, et al. Neuroprotection in ischemia: Blocking calcium-permeable acid-sensing ion channels. Cell 2004;118:687-98.
- Berdiev BK, Xia J, McLean LA, Markert JM, Gillespie GY, Mapstone TB, . Acid-sensing ion channels in malignant gliomas. J Biol Chem 2003; 278: 15 023–34.
- Vila-Carriles WH, Kovacs GG, Jovov B, Zhou ZH, Pahwa AK, Colby G, . Surface expression of ASIC2 inhibits the amiloride-sensitive current and migration of glioma cells. J Biol Chem 2006; 281: 19 220–32.
- Shen C, Buck AK, Liu X, Winkler M, Reske SN. Gene silencing by adenovirus-delivered siRNA. FEBS Lett 2003;539:111-4.
- Barton GM, Medzhitov R. Retroviral delivery of small interfering RNA into primary cells. Proc Natl Acad Sci USA 2002; 99: 14 943–5.
- Devroe E, Silver PA. Retrovirus-delivered siRNA. BMC Biotechnol 2002;2:15-9.
- Abbas-Terki T, Blanco-Bose W, Deglon N, Pralong W, Aebischer P. Lentiviral-mediated RNA interference. Hum Gene Ther 2002;13:2197-201.
- Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003;9:347-51.
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Biotechnol 2003;21:639-44.
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 2003;33:401-6.
- Zhang XC, Shan PY, Jiang DH, Noble PW, Abraham NG, Kappas A, . Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. J Biol Chem 2004; 279: 10 677–84.
