Rho GTPases of the RhoBTB subfamily and tumorigenesis1
Introduction
Since the late 1970s, when the first gene involved in tumor development in human was cloned, more than 200 tumor-related genes have been identified. They constitute a heterogeneous group of regulators of physiological processes that includes hormones, growth factors, receptors, cell adhesion molecules, signal transduction mediators, and transcription factors. Ras is the most widely studied oncogene in human carcinogenesis, and its discovery stimulated the search for Ras-related genes. Today, the Ras superfamily constitutes a numerous group of small guanosine triphosphatases (GTPases) that comprises over 150 members in humans, but can be found in all eukaryotes[1,2]. The common feature of Ras-related proteins is a ~20 kDa domain that, with few exceptions, binds and hydrolyzes GTP. Ras proteins act as molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state. Activation enables the GTPase to interact with a multitude of effectors that relay upstream signals to other components, eliciting diverse cellular responses. Two classes of molecules modulate the activation/inactivation cycle: guanine nucleotide exchange factors (GEF) and GTPase-activating proteins (GAP). In addition, guanine nucleotide-dissociation inhibitors regulate cycling of some GTPases between membranes and cytosol. The members of the Ras superfamily can be divided into several families based on sequence similarities, such as the extensively studied Ras, Rho, Rab, Arf, Ran, and Miro families[1,2], and broadly speaking, each family participates in the regulation of a major cellular process.
Rho GTPases are major regulators of the actin filament system and consequently of all processes that depend on the reorganization of the actin cytoskeleton, but they also participate in signaling pathways that regulate gene expression, cell cycle progression, apoptosis, and tumorigenesis[3–5]. Rho GTPases are being extensively studied in eukaryotes, from plants to mammals. In humans, the family comprises 21 members that have been grouped into subfamilies: Cdc42-like (Cdc42, TC10, TCL, Chp/Wrch-2, Wrch-1), Rac-like (Rac1–3, RhoG), Rho-like (RhoA–C), Rnd (Rnd1–2, Rnd3/RhoE), RhoD (RhoD and Rif), RhoH/TTF and RhoBTB (RhoBTB1–3)[4]. Although RhoBTB3 is frequently left outside because of its divergent GTPase domain, there is compelling architectural, phylogenetic, and possibly functional evidence for grouping this protein within the RhoBTB subfamily.
The RhoBTB subfamily constitutes the most recent addition to the Rho family. It was identified during the study of Rho-related protein-encoding genes in Dictyostelium discoideum[6]. Orthologs have been found in numerous eukaryote clades, but are absent in fungi, plants, and some metazoa[7]. RhoBTB proteins are remarkable for their unusual domain architecture: all RhoBTB proteins possess additional domains beyond the GTPase domain, in particular, a tandem of broad complex, tramtrack, bric à brac (BTB) domains (from the Drosophila transcription factors where the domain was first described) that explains the name given to the family and fully justifies their inclusion in the group of so-called atypical Rho GTPases[8].
Interest in the RhoBTB subfamily arose when RHOBTB2, the gene encoding the homonymous protein, was identified as the gene homozygously deleted in breast cancer samples and was proposed as a candidate tumor suppressor gene, dubbed DBC2 (deleted in breast cancer 2)[9]. The same property has been attributed recently to RHOBTB1[10]. RhoBTB proteins can be therefore incorporated into the group of Rho GTPases involved in tumorigenesis, although the mechanism RhoBTB proteins use differs radically from those of more typical Rho proteins[11] and may involve the direct targeting of other proteins for degradation in the 26S proteasome.
In this review we will summarize what we know about RhoBTB proteins, starting with basic aspects, such as domain architecture and gene expression, followed by the evidence that has accumulated during the last few years linking RhoBTB proteins with cancer. We will then connect this information with the roles that emerge from functional studies performed on the mammalian, and more limited, slime mold and Drosophila orthologs. We will close this review with an attempt to integrate all the available structural and functional information into a model that explains the participation of RhoBTB proteins in tumorigenesis.
Structure of RhoBTB proteins
The most salient feature of RhoBTB proteins is their domain architecture, which is, in general terms, shared by all members of the subfamily. In these proteins, the GTPase domain is followed by a proline-rich region, a tandem of 2 BTB domains, and a conserved C-terminal region (Figure 1). As already mentioned, in humans, the RhoBTB subfamily is composed of 3 isoforms: RhoBTB1, RhoBTB2, and RhoBTB3. RhoBTB1 and RhoBTB2 are very similar to each other and to the Drosophila ortholog (DmRhoBTB), whereas RhoBTB3 and the Dictyostelium discoideum ortholog (RacA) are the most divergent members. Here we will describe each domain and will discuss its functionality as well as some variations found in individual members.
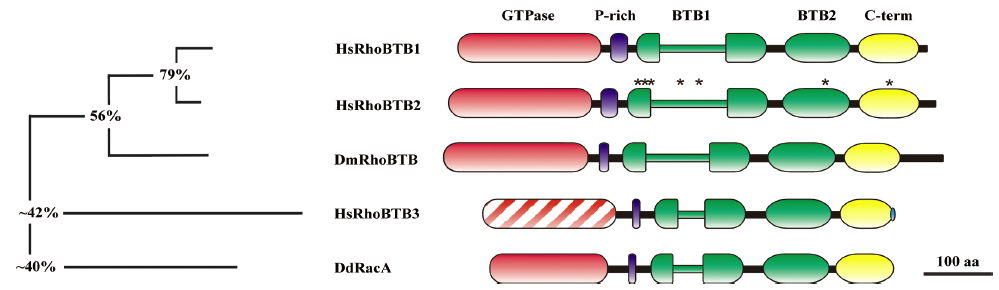
GTPase domain The GTPase domain is perhaps the region where most divergence is found among members of the RhoBTB subfamily. Early analyses revealed that this domain is typically Rac-like in RacA and divergent, but recognizable as Rho-related in RhoBTB1 and RhoBTB2 as well as in DmRhoBTB[6]. In RhoBTB3, the GTPase domain appears extensively erased, to the point that it is virtually unrecognizable as a GTPase. Only a short stretch at the end of the domain can be reliably aligned to the GTPase domain of other subfamily members. Consequently, RhoBTB3 does not bind GTP in vitro (Berthold J et al, personal communication). In phylogenetic analyses, the GTPase domain of RacA groups together with GTPases of the Rac subfamily and all relevant residues for nucleotide binding and enzymatic activity are conserved. In RacA the so-called Rho insert, a hypervariable insertion characteristic for Rho proteins, is shorter (6 amino acids) than the usual 13 amino acids of most Rac proteins. As far as it has been examined, the GTPase domain of RacA behaves like other Rac proteins (see functions of RhoBTB proteins below).
The GTPase domain of RhoBTB proteins other than RhoBTB3 and RacA also contains a Rho insert that is longer than usual (18 residues or more) and rich in charged residues. Moreover, the GTPase domain of these RhoBTB contains 2 insertions and 1 deletion, as well as a few deviations from the GTPase consensus of most Rho GTPases[6]. The insertions are placed immediately before (6 residues) and after (10 residues) the switch I. The deletion (2 residues) affects the phosphate/magnesium binding region 3 within the switch II; in particular, one of the deleted residues is the glutamine equivalent to Q61 in Ras. Also of importance, the glycine residue equivalent to G12 in Ras appears substituted by asparagine in RhoBTB1 and RhoBTB2 or threonine in DmRhoBTB. Because these 2 residues are essential for GTP hydrolysis, these proteins would predictably display impaired enzyme activity. Indeed, using a blot overlay approach, Chang and coworkers have shown that the GTPase domain of RhoBTB2 appears not to bind GTP at all, although this aspect requires biochemical confirmation[12].
Proline-rich region The proline-rich region links the GTPase to the first BTB domain. Sequences rich in proline are very common recognition motifs involved in protein–protein interactions. Among the modules that bind proline-rich regions are the SH3 (Src homology 3) domain, the WW domain, the Ena/VASP homology 1 domain, profilin, the GYF domain, ubiquitin enzyme variant (UEV), and the cytoskeleton-associated protein glycine-rich domain[13,14]. The proline-rich region of some RhoBTB proteins could act as a SH3 domain-binding site. The SH3 domain is often present in proteins involved in signal transduction and cytoskeleton organization. The PxxP motif (where x denotes any amino acid), initially described as the core binding motif of the SH3 domain, can be found in RhoBTB1, RhoBTB2, and DmRhoBTB, where the proline-rich region is prominent, and in RacA, but not in RhoBTB3 where this region is very poorly preserved. Subsequent analyses have defined proline-rich motifs for a number of different SH3 domains more precisely as +xΦPxΦP (class I ligands) and ΦPxΦPx+ (class II ligands; where Φ is a hydrophobic and + is in most cases a basic residue)[13,15]. Interestingly, RhoBTB1 and RhoBTB2 have a conserved class II motif. DmRhoBTB has a motif that matches the more recently recognized class III ligands with the (R/K)xx(K/R) sequence[14]. Nevertheless, albeit the sequence analysis strongly suggests that the proline-rich region of several RhoBTB proteins is a potential SH3 domain-binding site, this still needs to be verified experimentally.
BTB domain The BTB domain, also known as a poxvirus and zinc finger domain, is an evolutionary conserved domain that is widespread among eukaryotes. In humans, nearly 200 different proteins bear BTB domains, in most cases, in combination with other domains. Two of the accompanying domains are particularly frequent, the zinc finger (ZF) and the Kelch domain. BTB–ZF proteins constitute a large family of transcription factors, whereas BTB–Kelch proteins play roles in the dynamics of the actin cytoskeleton[16,17].
The BTB domain has been known for long time as a protein–protein interaction domain participating in homomeric and heteromeric associations with other BTB domains[18]. More recently, a series of papers almost simultaneously identified this domain as a component of cullin3-dependent ubiquitin ligase complexes[19–22]. These complexes constitute a class of the very large family of ubiquitin ligases[23], which catalyze the addition of ubiquitin, a highly conserved 76-amino acid globular protein, to a number of target proteins. This post-translational modification labels proteins for degradation by the 26S proteasome, although other cellular functions not directly involving protein degradation are also controlled by this modification[24].
Cullins (of which there are 7 in mammals) function as scaffolding proteins that bring together the ubiquitin-conjugating enzyme and substrate-recognition components. The core ligase of a cullin-dependent complex consists of a cullin protein that binds through its C-terminus the RING-finger protein Roc1 (which recruits the ubiquitin-conjugating enzyme) and through its N-terminus, a linker protein. An adaptor protein then acts as a bridge between the linker protein and the substrates. The complex is positively regulated by covalent attachment of the Nedd8 ubiquitin-like protein to the cullin subunit. Each cullin family member interacts with a specific adaptor. The cullin1 and cullin7 complexes contain the Skp1 linker and an F-box-containing adaptor, whereas the cullin2 and cullin5 complexes contain the linker elongin C (along with elongin B) and a SOCS-box-containing protein. The cullin3 complexes contain a BTB domain-bearing protein that interestingly functions simultaneously as a linker and adaptor[25].
Closer inspection of the structure of the BTB domain in comparison with that of Skp1 and elongin C revealed a similar folding, and predictably a common interface for interaction with the corresponding cullin, despite a low degree of primary sequence conservation. In fact, elongin C and Skp1 are now considered BTB proteins. The common folding of all these proteins consists of a 95 amino acid globular cluster of 5 α-helices flanked by 3 short β-strands. The BTB domains of the BTB–ZF, BTB–Kelch, and RhoBTB proteins contain an N-terminal extension that folds into 1 α-helix and 1 β-strand, and this extension mediates the formation of dimers and oligomers[17].
The BTB domains of RhoBTB have some special features. A tandem of 2 BTB domains as in RhoBTB is not frequently found within the BTB protein family. Moreover, the first BTB domain is bipartite, being interrupted by an extension of unknown function that varies in length and composition among RhoBTB proteins. In RhoBTB1, RhoBTB2, and DmRhoBTB the insertion is 3 times longer (up to 100 residues) than in RhoBTB3 and RacA, and is in all cases rich in charged residues. Because the BTB domains of RhoBTB are of the extended type, these proteins are predicted to exist as dimers, and in fact, they are capable of forming homodimers and heterodimers (Berthold J et al, personal communication). The role of RhoBTB as components of the cullin3-dependent complexes will be discussed below.
The C-terminal region Following the second BTB domain, there is a region conserved in all members of the RhoBTB subfamily that may constitute a novel domain, but has not been found so far in any other protein apart from RhoBTB. The core of the C-terminal domain consists of approximately 80 amino acids that predictably folds as 4 consecutive α-helices. The last helix ends close before the prenylation signal of RhoBTB3, but prolongs further in a predicted β-strand in RhoBTB1, RhoBTB2, and DmRhoBTB[7].
Although Rho GTPases typically bear a CAAX motif, only RhoBTB3 conserves this feature. This motif is recognized by a set of enzymes that introduce a post-translational modification, isoprenylation, responsible for the targeting of the modified protein to membranes. Closely upstream of this motif there is an additional cysteine residue in RhoBTB3, which suggest that this protein might also be palmitoylated[7]. The presence of nuclear localization signals in the C-terminus of some members of the RhoBTB subfamily has been occasionally reported[8,12], but is controversial because computer programs commonly used to predict these signals often yield inconsistent results. Unlike for many BTB proteins that function as transcription factors, there is no experimental evidence showing the nuclear localization of RhoBTB.
Expression of RHOBTB genes
Both in humans and mice, all 3 RHOBTB genes are rather ubiquitously expressed, although with notable differences in the pattern of tissue levels among the 3 genes. In humans, where expression has been studied using multiple-tissue Northern dot blots and quantitative PCR[7,26,27], RHOBTB1 showed high levels in skeletal muscle and placenta followed by the stomach, kidney, testis, adrenal gland, and uterus, whereas RHOBTB3 is highly expressed in the placenta, testis, pancreas, adrenal and salivary glands, and neural and cardiac tissues. RHOBTB2 is very weakly expressed, but relatively high levels were detected in neural and cardiac tissues. All 3 genes are expressed in fetal tissues.
The expression pattern of the mouse counterparts has been analyzed in conventional Northern blots and is roughly comparable to that of the human genes[7]. Mouse RHOBTB1 is highly expressed in the heart, testis, and kidney, and moderately in the uterus, liver, lung, stomach, placenta, and skeletal muscle. Mouse RHOBTB3 is strongly expressed in the brain, heart, and uterus, and moderately in all other tissues. As in human tissues, RHOBTB2 is very weakly expressed in mouse tissues, with relatively higher expression levels in the brain. In addition, the expression of 1 or more RHOBTB genes has been reported in numerous human and mouse cell lines using RT–PCR. In Northern blot analyses, mouse RHOBTB3 and RHOBTB1 appear as single 5 kb transcripts, although in most tissues, a less prominent 4 kb RHOBTB1 transcript is also expressed. RHOBTB2 is equally expressed both as 4 kb and 5 kb transcripts. The 2 transcripts in these genes have been explained by the use of alternative promoters or by alternative splicing in the 5´UTR, but this issue has not been addressed and remains speculative[7].
RHOBTB3 has been reported in RNA from whole mouse embryos in Northern blot analyses, where a transcript was detected from embryonic d 11.5, declining at d 17.5[7]. RHOBTB2 has been reported in several fetal tissues using RT–PCR[28]. Using in situ hybridization, the high and specific expression of RHOBTB2 has been observed in the central and peripheral nervous system and comparatively weaker in the gut during mouse embryogenesis, but the mRNA becomes undetectable at embryonic d 18.5[28]. Although still limited, these data implicate RHOBTB genes in controlling developmental processes.
With RHOBTB2 having been described as a tumor suppressor gene involved in breast cancer, it was of interest studying the expression of this gene during mammogenesis. Using RT–PCR and Northern blot analysis, St-Pierre et al[28] found that during mammary gland development in mice, RHOBTB2 transcripts are expressed at low but constant levels. However, attempts to study the spatial pattern of the expression of RHOBTB2 in the mammary gland using in situ hybridization were inconclusive because of undetectable mRNA levels. Our own attempts to study the expression of RHOBTB genes at the cellular level using in situ hybridization on adult mouse tissues were hampered by the very low mRNA levels of these genes. While the expression of RHOBTB2 was undetectable in all of the tissues analyzed, RHOBTB1 and RHOBTB3 mRNA were found in the endothelial cells of the heart as well as in spermatocytes and spermatides in the testis. Additionally, RHOBTB1 and RHOBTB3 messages were detected in large vessels of the kidney and brain, respectively (Berthold J et al, personal communication).
RhoBTB in cancer
Since the first report proposing RHOBTB2 as a tumor suppressor gene, evidence is accumulating in support of members of the RhoBTB subfamily being implicated in tumorigenesis (Table 1). The RHOBTB2 gene was identified as the gene homozygously deleted at region 8p21 in breast cancer samples[9]. This is a region commonly associated with loss of heterozygosity (LOH) in a wide range of cancers. Hamaguchi and coworkers performed a representational deletion analysis on a large sample of breast tumors using DNA markers for the 8p21 region and found that RHOBTB2 was homozygously deleted in 3.5% of the tumors. A mutation analysis revealed 2 somatic missense mutations in breast tumors and 2 more missense mutations each in a breast and a lung tumor cell line. The expression of RHOBTB2 appeared extinguished in approximately 42% of breast and 50% of lung cancer cell lines[9]. A more extensive mutation analysis of breast cancers revealed some polymorphisms as well as 2 novel somatic mutations in the promoter and 5´UTR of RHOBTB2 in sporadic tumors, but no additional mutations in the coding region of sporadic or familial cancers[29].

Full table
In a study addressing RHOBT2 in bladder cancer, Knowles et al performed a LOH and mutation analysis on tumor samples and cell lines[30]. They found LOH in the target region in 42% of informative tumors. A sequence analysis revealed numerous polymorphisms and 1 missense somatic mutation. In addition, the expression of RHOBTB2 was found to be reduced by 2 to 20-fold in 9 of 12 cell lines with predicted LOH in the region of interest. In a study on primary gastric cancers, LOH was found in 29% of tumors; a sequence analysis identified several polymorphisms and 1 more missense somatic mutation[31].
In a recent study on head and neck cancer, RHOBTB1 has also been postulated as a tumor suppressor gene[10]. The 10q21 region where the RHOBTB1 gene is located has been identified as a hotspot region in head and neck squamous cell carcinomas (HNSCC) in a genome-wide LOH analysis[32]. Focusing on RHOBTB1, Beder et al found a high frequency of LOH with a microsatellite marker located in intron 7 of the gene[10]. In 12 of 52 tumor samples, LOH could be demonstrated, and interestingly, 4 samples showed LOH exclusively for the RHOBTB1 locus. Since almost 50% of the tumor samples were not informative in the LOH analysis, it is very likely that the RHOBTB1 locus is affected in a higher proportion of tumors. A mutation analysis revealed 3 polymorphisms, but no pathogenic mutations. The expression of RHOBTB1 decreased in 37% of the samples analyzed, although it increased in 35%, but significantly, all low-expression samples for which informative allelic loss data were available displayed LOH.
Although we still have limited information on the status of RHOBTB genes in tumors, the picture that emerges from the reports discussed above is one of rare mutations but common reduced or extinguished expression. This observation can be made extensive to the third family member, RHOBTB3. We have determined the expression of RHOBTB3 in an array of tumor tissues and their matched normal tissues and have found a moderate but significant decrease of RHOBTB3 expression in the breast, kidney, uterus, lung, and ovary tumors (Berthold J et al, personal communication). It appears that mechanisms other than mutations are more frequently implicated in the inactivation of these genes. One such mechanism may be promoter methylation. The hypermethylation of CpG islands results in the downregulation or complete abrogation of gene expression and is a frequent epigenetic alteration in primary tumors[33]. The promoter region of RHOBTB2 has a CpG island, and in RHOBTB1, the promoter region and exon 1 (an untranslated exon) have a high GC content and numerous CpG motifs. Interestingly, the mutations found in the promoter and 5´UTR of RHOBTB2 in some breast tumors might affect the regulation of gene expression[29]. The –238G>A polymorphism abolishes a putative Sp1-1 binding site and creates an additional CpG dinucleotide. The –121C>T mutation abolishes binding sites for the transcription factors E2F and snail, and the +48G>A mutation creates a putative binding site for the bZIP910 transcription factor. Clearly, future work should be directed to analyze this aspect of RHOBTB gene expression in tumor tissues and cell lines.
Functions of RhoBTB
The role of RHOBTB genes as tumor suppressors, initially attributed to RHOBTB2, more recently to RHOBTB1, and probably extensively also to RHOBTB3, is receiving increasing support. Nevertheless, the mechanisms by which RhoBTB proteins exert this and other roles remain largely speculative. Siripurapu et al have taken a large scale approach to explore the roles of RHOBTB2[34]. They constitutively expressed RHOBTB2 in HeLa cells, followed by silencing of the ectopic gene and then a microarray analysis. A comparison of the overexpressing and silenced samples revealed significant alterations in genes belonging to 2 networks: one that regulates cell growth through cell cycle control and apoptosis and one that is related to cytoskeleton and membrane trafficking. Although the approach used in this study is adventurous and rather artificial, evidence is accumulating in support of the roles for RhoBTB proteins in the processes revealed by Siripurapu et al[34]. The identification of RhoBTB2 as a component and substrate of cullin3-dependent ubiquitin ligase complexes was key for the mechanistic understanding of RhoBTB functioning[35]. Although several potential roles of RhoBTB proteins are considered separately, they are probably interrelated. It is also very likely that the function as adaptors of cullin3-dependent ubiquitin ligases constitutes the underlying mechanism for all other roles, therefore, it will be discussed first and more extensively.
RhoBTB as adaptors of cullin3-dependent ubiquitin ligases The identification of the BTB domain as adaptor in cullin3-dependent ubiquitin ligase complexes prompted Wilkins and coworkers to investigate whether RhoBTB2 may also take part in the formation of such complexes[35]. They identified the N-terminal region of murine cullin3 as an interacting partner of RhoBTB2 in a yeast 2 hybrid screening[35]. RhoBTB2 interacts specifically with cullin3, but not other cullin family members in vivo; the interaction was mapped to the first BTB domain in a series of pull-down experiments with deletion constructs. Wilkins et al also provided evidence that RhoBTB2 is itself a substrate for cullin3-based ubiquitin ligase complexes, as treatment with proteasomal inhibitor MG132 or shRNA ablation of cullin3 resulted in increased levels of RhoBTB2, and RhoBTB2 was polyubi-quitinylated by cullin3 complexes in vitro[35]. Our own unpublished data indicate that many of these properties are shared by all members of the RhoBTB subfamily, including RhoBTB3.
RHOBTB2 was proposed as a candidate tumor suppressor gene based on the fact that its re-expression in T-47D (a breast cancer cell line that lacks RHOBTB2 transcripts) caused growth inhibition, whereas the expression of the somatic mutant D299N did not have the same effect[9]. This mutation is placed in the first BTB domain immediately before the insertion. In fact, it is interesting that almost all missense mutations found in the RHOBTB2 locus reside in the first BTB domain of the protein (Figure 1). The question arises whether one or more of those mutations result in impaired interaction with cullin3. This has been investigated by Wilkins and coworkers who found that the Y284D mutant, but not the D299N and D368A mutants, failed to coimmuno-precipitate with cullin3, and consequently, had a longer half-life than the wild-type protein[35]. The Y284D mutation resides in the dimerization interface of the first BTB domain and could prevent proper folding. Analogous mutants have been shown to abrogate function by impairing folding of the BTB domain, for example, in the transcription factor PLZF[36].
We have found a correlation in the expression changes between RHOBTB3 or RHOBTB1 and CUL3 in tumor tissues (Berthold J et al, personal communication), supporting the view that RHOBTB genes and CUL3 may be coregulated and the role of RhoBTB in tumorigenesis is related to its role as adaptor for cullin3-dependent ubiquitin ligase complexes. There are approximately 200 genes encoding BTB proteins in the human genome, suggesting that a significant proportion of cullin3-dependent complexes might control the ubiquitinylation and degradation of cancer-related proteins through multiple mechanisms. In fact, several BTB proteins have been found to be linked to tumorigenesis, although their roles in the formation of cullin3-dependent complexes have generally not been addressed. To cite a few examples, the tumor suppressor gene HIC1 (hypermethylated in cancer 1) is located at a region of chromosome 17 that is frequently hypermethylated or deleted in human tumors. It works as a transcriptional repressor functionally cooperating with p53 to suppress the age-dependent development of cancer[37]. The transcriptional repressor and candidate oncogene Bcl-6 is an important regulator of lymphoid development and function. The BCL6 gene is localized in a region implicated in chromosomal translocations frequently found in non-Hodgkin’s lymphoma of B-cell type[38]. The Kelch-related Mayven has been proposed to promote tumor growth through the induction of c-Jun and cyclin D1[39]. Another example is Kaiso, involved in p120-catenin/Kaiso signaling pathways that regulate gene expression in development and carcinogenesis[40]. In fact, the role of cullin3-dependent complexes in tumorigenesis can be placed in the wider context of the cullin family where every member has been found to be implicated in ubiquitinylation of cancer-related substrates (see Guardavaccaro and Pagano[41] for a comprehensive review).
RhoBTB, cell growth, and apoptosis As already mentioned, Hamaguchi and coworkers reported that the overexpression of RhoBTB2 in the breast cancer cell line T-47D effectively suppressed cell growth in vitro[9]. More recently, Freeman and coworkers have shown that the overexpression of RhoBTB2 leads to a short-term increase in cell cycle progression and proliferation, but long-term expression has a negative effect on proliferation[42]. The growth arrest effect of RhoBTB2 has been explained by the downregulation of cyclin D1. Cyclin D1 is upstream of cyclin E, and the overexpression of any of both prevented the growth arrest effect of RhoBTB2[43]. The effect on cyclin D1 is probably post-transcriptional, but only partially dependent on proteasomal degradation. Moreover, it has not been investigated whether cyclin D1 is degraded by cullin3-dependent complexes through direct binding to RhoBTB2. In this respect it is important to note that one mechanism as to how cullin3-dependent complexes regulate the cell cycle is through the targeting of cyclin E for ubiquitinylation[44]. The downregulation of cyclin D1 is essential for the cell proliferation suppression effect of RhoBTB2, but this works for T-47D cells and not for 293 cells. It therefore appears that the regulation of cyclin D1 is not a universal tumor suppressive mechanism used by RhoBTB2. The explanation has been put forward that resistance to RhoBTB2 in some cell lines may be achieved by rapid destruction of the protein through 26S proteasome-mediated degradation[45]. Further support for the roles in cell cycle regulation has been provided recently with the identification of RHOBTB2 as a target of the E2F1 transcription factor[42]. E2F1 is a member of a class of E2F implicated in the transcription of genes necessary for DNA replication and cell cycle progression and can also promote apoptosis[46]. RhoBTB2 levels increase upon initiation of prophase and decrease at telophase, and this effect depends on E2F1[42].
RhoBTB2 levels also increase during drug-induced apoptosis in an E2F1-dependent manner, and the downregu-lation of RHOBTB2 delays the onset of apoptosis[42]. In agreement with an implication in this process, RhoBTB was found in Drosophila as one of several genes whose expression was significantly upregulated in a DNA microarray analysis aimed at identifying genes associated with cell death induced by the steroid hormone ecdysone[47]. Interestingly, in this study, additional genes encoding Rho-signaling components, most notably Rac2, also appeared upregulated. However, the role of RhoBTB as a candidate cell death regulator was not investigated further.
RhoBTB and vesicle transport Chang et al have addressed the potential role of RhoBTB2 in vesicle transport in a fluorescent recovery after photobleaching analysis with the help of a vesicular stomatitis virus glycoprotein (VSVG) fused to GFP[12]. VSVG is extensively used to study anterograde transport from the endoplasmic reticulum to the Golgi apparatus. Knockdown of endogenous RhoBTB2 hindered the ER to Golgi apparatus transport and resulted in the altered distribution of the fusion protein. In this study, the authors found that GFP–RhoBTB2 was distributed in a vesicular pattern when expressed at low levels. Some of the vesicles appeared adjacent to microtubules and an intact microtubule network seemed to be required for the mobility of RhoBTB.
The localization of RhoBTB1 and RhoBTB2 in vesicular structures had been postulated before. Aspenström and coworkers reported the accumulation of the ectopically-expressed proteins at perinuclear structures that did not colocalize with lysosomal or Golgi apparatus markers[48]. These structures apparently represent aggregates, and can be also induced upon the ectopic expression of RhoBTB3. However, when RhoBTB3 is expressed at moderate levels, it displays a vesicular pattern. Many of the vesicles colocalize with early endosome markers, and localization in close vicinity of microtubules is also apparent. As mentioned earlier, RhoBTB3 ends with a prenylation motif, and the C-terminal extension of RhoBTB3 is necessary and sufficient for the attaching of the protein to vesicles (Berthold J et al, personal communication). However, prenylation might not be the only mechanism required for the targeting of RhoBTB to vesicles as RhoBTB1 and RhoBTB2 lack a prenylation motif.
Further, in support of a role in vesicle trafficking, RhoBTB has been identified as one of the genes that suppress the neuromuscular junction overgrowth phenotype induced in Drosophila larvae by the expression of a dominant negative form of the N-ethylmaleimide sensitive factor (NSF)[49]. NSF is an ATPase that participates in vesicle trafficking through binding to the SNARE complex and is also important for the regulation of receptor trafficking[50]. Interestingly, NSF is one gene whose expression appeared altered in the study of Siripurapu et al discussed earlier[34], which is suggestive of a conserved mechanism that requires further investigation. If a role for these proteins in vesicle trafficking gains support, then RhoBTB will engross the growing list of Rho GTPases involved in this process. The mechanism remains obscure, but will be most likely an unusual one.
RhoBTB and the actin filament system Although very atypical, RhoBTB proteins are members of the Rho family, therefore, the first aspect that was investigated was their effect on the organization of the actin filament system. Aspenström and coworkers observed a moderate influence, if at all, on the morphology and actin organization of porcine aortic endothelial cells upon the ectopic expression of RhoBTB1 and RhoBTB2[48], an observation that we have made extensive to RhoBTB3 and several other cell lines (Berthold J et al, personal communication). Not surprisingly, neither RhoBTB1 nor RhoBTB2 were found to interact with the GTPase-binding domain of WASP, PAK1, or Rhotekin, 3 well-known effectors of many typical Rho GTPases[48]. Confirming that, at least in metazoa, RhoBTB proteins do not play a major role in the organization of the actin filament system, DmRhoBTB was found among the proteins whose depletion had no effect on lamellae morphology in Drosophila S2 cells[51]. These cells can be induced to spread when plated on a concanavalin A-coated surface and constitute then an appropriate system to study the formation of lamellae.
Unlike metazoan RhoBTB, the Dictyostelium ortholog RacA may be directly implicated in the regulation of the actin cytoskeleton, although the evidence is indirect and no functional studies have been published yet. The racA gene is very weakly expressed throughout the life cycle of Dictyostelium[6], but the protein is present at all stages. The GTPase domain of RacA, which as already mentioned, is very closely related to members of the Rac subfamily, is able to interact with the Rac-binding domain of WASP and kinases of the PAK family in yeast 2 hybrid assays[52–54], although these interactions remain to be demonstrated in vivo. Unlike metazoan RhoBTB, RacA is susceptible to regulation by RhoGEF and RhoGAP, and in vitro interaction with a RhoGEF, GxcDD, has been reported recently[55]. We speculate that RacA represents a “primitive” cytoskeleton-regulating stage of the RhoBTB subfamily that was replaced in the evolved metazoan RhoBTB proteins by roles in cell proliferation and vesicle trafficking.
Conclusion
There is increasing evidence linking Rho-regulated signal transduction pathways to tumorigenesis and metastasis[11,56,57]. Rho GTPases play a role in the acquisition of an invasive phenotype of tumor cells, either directly via their effects on the cytoskeleton, or indirectly via changes in gene transcription. It is noteworthy that with the exception of RacH and perhaps Rac1, no mutations in typical Rho GTPases have been found to be associated specifically with tumors. It is rather alterations in the expression or activation levels of these proteins which characterizes many tumors. For example, the expression of Rac1b (an alternative splice variant of Rac1) increases in colorectal tumors[58], the overexpression of RhoC correlates with the invasiveness of non-small cell lung cancer[59], and Cdc42 is overexpressed in HNSCC[60].
Unlike typical Rho GTPases, those of the RhoBTB subfamily appear to play a part in the carcinogenic process through a mechanism that involves the downregulation or loss of function. Taking into consideration the ability of RhoBTB proteins to constitute cullin3-dependent complexes, a model emerges in which these proteins recruit substrates for degradation in the 26S proteasome (Figure 2). While the first BTB domain is involved in recruitment of cullin3 and associated components, other regions of the protein, such as the GTPase domain and the C-terminal conserved region or even the second BTB domain, would function as substrate recognition domains. The insertion of the first BTB domain probably folds away from the globular BTB core and might also be implicated in substrate recognition, whereas the proline-rich region of RhoBTB1 and RhoBTB2 could play regulatory roles through interaction with SH3 domain-bearing proteins.
RhoBTB proteins would be required to maintain constant levels of putative substrates, thus exerting regulatory roles during the cell cycle, vesicle transport, and in lower eukaryotes, cytoskeleton homeostasis. It is easy to understand that situations that result in the impaired expression of RHOBTB genes, or more rarely, mutations that result in impaired functioning (binding to cullin3, dimerization, interaction with substrates, targeting) of the protein might lead to the accumulation of RhoBTB substrates and alterations of the cellular homeostasis. Such a regulatory mechanism could be the basis of the tumor suppressor role of RhoBTB proteins and is analogous to the well-studied role of the von Hippel–Lindau (VHL) tumor suppressor. VHL is an adaptor for cullin2-dependent ubiquitin ligase complexes that target the hypoxia-inducible factor for degradation. Nearly 70% of naturally-occurring cancer-predisposing mutations of VHL disrupt the formation of these complexes[61]. Obviously, if we wish to clear the mechanisms as to how the malfunction of RhoBTB proteins results in tumor formation, we imperatively need to know how these proteins are regulated at all levels (transcriptional, translational, and post-translational) and what their substrates are.

References
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005;118:843-6.
- Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE 2004; 2004: RE13.
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005;21:247-69.
- Wennerberg K, Der CJ. Rho family GTPases: it’s not only Rac and Rho (and I like it). J Cell Sci 2005;117:1301-12.
- Burridge K, Wennerberg K. Rho and Rac take center stage. Cell 2004;116:167-79.
- Rivero F, Dislich H, Glockner G, Noegel AA. The Dictyostelium discoideum family of Rho-related proteins. Nucleic Acids Res 2001;29:1068-79.
- Ramos S, Khademi F, Somesh BP, Rivero F. Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene 2002;298:147-57.
- Aspenström P, Ruusala A, Pacholsky D. Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp Cell Res 2007;313:3673-9.
- Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, et al. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci USA 2002;99:13647-52.
- Beder LB, Gunduz M, Ouchida M, Gunduz E, Sakai A, Fukushima K, et al. Identification of a candidate tumor suppressor gene RHOBTB1 located at a novel allelic loss region 10q21 in head and neck cancer. J Cancer Res Clin Oncol 2006;132:19-27.
- Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays 2005;27:602-13.
- Chang FK, Sato N, Kobayashi-Simorowski N, Yoshihara T, Meth JL, Hamaguchi M. DBC2 is essential for transporting vesicular stomatitis virus glycoprotein. J Mol Biol 2006;364:302-8.
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 2000;14:231-41.
- Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J 2005;390:641-53.
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci 2001;114:1253-63.
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein–protein interaction domain. Bioessays 2006;28:1194-202.
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol 2005;6:R82.
- Aravind L, Koonin EV. Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J Mol Biol 1999;285:1353-61.
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 2003;425:311-6.
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 2003;12:783-90.
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 2003;5:1001-7.
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003;425:316-21.
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002;82:373-428.
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2001;2:169-78.
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005;6:9-20.
- Nagase T, Ishikawa KI, Suyama M, Kikuno R, Hirosawa M, Miyajima N, et al. Prediction of the coding sequences of unidentified human genes. XII. The complete sequence of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 1998;5:355-64.
- Nagase T, Ishikawa KI, Suyama M, Kikuno R, Miyajima N, Kotani H, et al. Prediction of the coding sequences of unidentified human genes. XI. The complete sequence of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 1998;5:277-86.
- St-Pierre B, Jiang Z, Egan SE, Zacksenhaus E. High expression during neurogenesis but not mammogenesis of a murine homologue of the deleted in breast cancer2/Rhobtb2 tumor suppressor. Gene Expr Patterns 2004;5:245-51.
- Ohadi M, Totonchi M, Maguire P, Lindblom A, Habibi R, Afshin Alavi B, et al. Mutation analysis of the DBC2 gene in sporadic and familial breast cancer. Acta Oncol 2007;46:770-2.
- Knowles MA, Aveyard JS, Taylor CF, Harnden P, Bass S. Mutation analysis of the 8p candidate tumour suppressor genes DBC2 (RHOBTB2) and LZTS1 in bladder cancer. Cancer Lett 2005;225:121-30.
- Cho YG, Choi BJ, Song JH, Zhang C, Nam SW, Lee JY, et al. Genetic analysis of the DBC2 gene in gastric cancer. Acta Oncol 2007. [Epub ahead of print].
- Beder LB, Gunduz M, Ouchida M, Gunduz E, Ito S, Sakai A, et al. Genome-wide analyses of loss of heterozygossity in head and neck squamous cell carcinomas. Lab Invest 2003;83:99-105.
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28.
- Siripurapu V, Meth J, Kobayashi N, Hamaguchi M. DBC2 significantly influences cell-cycle, apoptosis, cytoskeleton and membrane-trafficking pathways. J Mol Biol 2005;346:83-9.
- Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev 2004;18:856-61.
- Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, et al. In-depth mutational analysis of the promyelocytic leukemia zink finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol 2000;20:6550-67.
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005;123:437-48.
- Ohno H. Pathogenetic role of BCL6 translocation in B-cell non-Hodgkin’s lymphoma. Histol Histopathol 2004;19:637-50.
- Bu X, Avraham HK, Li X, Lim B, Jiang S, Fu Y, et al. Mayven induces c-Jun expression and cyclin D1 activation in breast cancer cells. Oncogene 2005;24:2398-409.
- van Roy F, McCrea P. A role for Kaiso-p120ctn complexes in cancer? Nat Rev Cancer 2005;5:956-64.
- Guardavaccaro D, Pagano M. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene 2004;23:2037-49.
- Freeman SN, Ma Y, Cress WD. Rhobtb2 (DBC2) is a mitotic E2F1 target gene with a novel role in apoptosis. J Biol Chem 2007. [Epub ahead of print].
- Yoshihara T, Collado D, Hamaguchi M. Cyclin D1 downregulation is essential for DBC2’s tumor suppressor function. Biochem Biophys Res Commun 2007;358:1076-9.
- Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 1999;13:2375-87.
- Collado D, Yoshihara T, Hamaguchi M. DBC2 resistance is achieved by enhancing 26S proteasome-mediated protein degradation. Biochem Biophys Res Commun 2007;360:600-3.
- DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med 2006;6:739-48.
- Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol 2003;13:350-7.
- Aspenström P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J 2004;377:327-37.
- Laviolette MJ, Nunes P, Peyre JB, Aigaki T, Stewart BA. A genetic screen for suppressors of Drosophila NSF2 neuromuscular junction overgrowth. Genetics 2005;170:779-92.
- Zhao C, Slevin JT, Whiteheart SW. Cellular functions of NSF: not just SNAPs and SNAREs. FEBS Lett 2007;581:2140-9.
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol 2003;162:1079-88.
- Han JW, Leeper L, Rivero F, Chung CY. Role of RacC for the regulation of WASP and phosphatidylinositol 3-kinase during chemotaxis of J Biol Chem 2006; 281: 35 224–34.
- Park KC, Rivero F, Meili R, Lee S, Apone F, Firtel RA. Rac regulation of chemotaxis and morphogenesis. EMBO J 2004;23:4177-89.
- de la Roche M, Mahasneh A, Lee SF, Rivero F, Coté GP. Cellular distribution and functions of wild-type and constitutively activated Dictyostelium PakB. Mol Biol Cell 2005;16:238-47.
- Mondal S, Neelamegan D, Rivero F, Noegel AA. GxcDD, a putative RacGEF, is involved in Dictyostelium development. BMC Cell Biol 2007;8:23.
- Aznar S, Lacal JC. Rho signals to cell growth and apoptosis. Cancer Lett 2001;165:1-10.
- Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer 2002;2:133-42.
- Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 1999;18:6835-9.
- Shikada Y, Yoshino I, Okamoto T, Fukuyama S, Kameyama T, Maehara Y. Higher expression of RhoC is related to invasiveness in non-small cell lung carcinoma. Clin Cancer Res 2003;9:5282-6.
- Abraham MT, Kuriakose MA, Sacks PG, Yee H, Chriboga L, Bearer EL, et al. Motility-related proteins as markers for head and neck squamous cell cancer. Laryngoscope 2001;111:1285-9.
- Maynard MA, Ohh M. The role of hypoxia-inducible factors in cancer. Cell Mol Life Sci 2007;64:2170-80.
