Affinity of anticancer drug, daunomycin, to core histones in solution: comparison of free and cross–linked proteins
Introduction
Daunomycin is an anthracycline antibiotic widely used in the treatment of myelogenous leukemia and solid tumors[1,2]. Numerous studies have revealed that nuclear DNA is an important target for this drug. The structure of daunomycin consists of 2 distinct domains (Figure 1): a planar aglycon chromophore that intercalates between adjacent base pairs of DNA and an amino sugar ring that lies in the minor groove of the DNA double helix[3,4]. Binding of daunomycin to DNA results in the inhibition of both, DNA replication and RNA transcription[5–7].
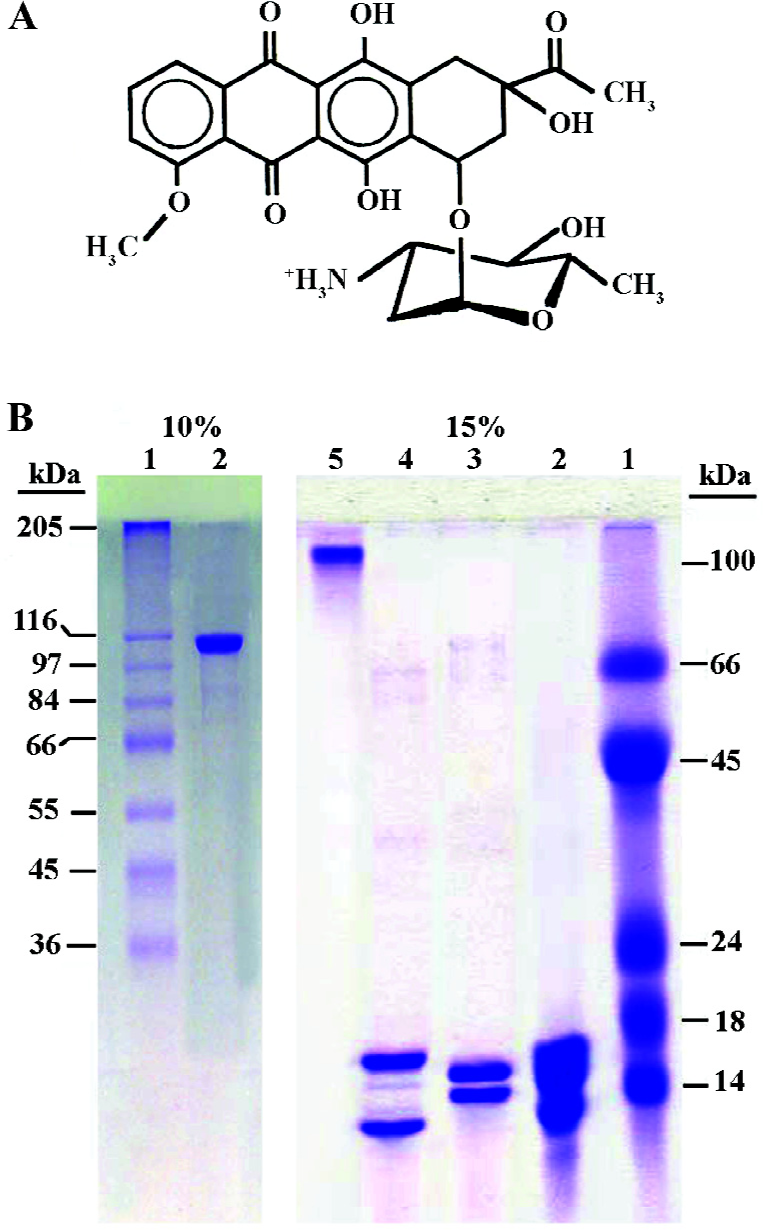
In the cell nucleus, DNA is compacted into chromatin, which is a complex structure built from repeating units (nucleosomes)[8]. These consist of 145 bp DNA wrapped around an octamer of basic proteins (core histones). The core histones are small, basic proteins ranging between 11 and 16 kDa, with more that 20% of their amino acids composition being lysine and argenines. There are 5 main histones: the linker histones of the H1 family and the 4 core or nucleosomal histones H2A, H2B, H3, and H4, which associate as H2A/H2B dimers and a H3/H4 tetramer, arranged in a octamer form: the DNA is wrapped within the nucleosome-chromatin unit[9].
How the occurrence of histones that bind to DNA affects the binding of daunomycin, is an important question when trying to understand the mechanisms of action of this drug at the chromatin level. To explore this question, some authors have focused on the binding of daunomycin to chromatin and nucleosomes and have shown that the binding affinity of daunomycin to chromatin increases upon the removal of histones[10–12]. We recently focused on the interaction of daunomycon with the histone H1 family (linker histones) in chromatin and in solution, providing evidence that the binding is cooperative and stabilizes the protein against thermal denaturation[13–15].
In the present study, we have extended our research on the interaction of daunomycin with the core histones free in solution and cross-linking with a bifunctional reagent to produce a complex designated, cross linked core (CL-core) resembling an octamer in nucleosomes. The results provide an evidence that daunomycin binds to free core histones to a greater extent than to the CL-core complex, implying that the environment of core histones in the chromatin plays a fundamental role in the drug-histone interaction.
Materials and methods
Chemicals Daunomycin hydrochloride (Figure 1A) was purchased from Sigma (St Louis, MO, USA). A stock solution of drug was prepared in sterile distilled water at a concentration of 2 mg/mL and stored at -20 ºC until further use. A dilution of the drug stock in the appropriate buffer was prepared immediately before use. The concentration of daunomycin was determined spectrophotometrically using an extinction coefficient of 11500 mol-1 cm-1 at 480 nm.
Preparation of histones The total histone containing all 5 histones was extracted from calf thymus by 0.25 mol/L HCl described by Johns[16]; the core histones were further purified using CM-Sephadex-C25 ion exchange chromatography (Pharmacia)[17]. The purified core histone was dissolved in 20 mmol/L Tris buffer (pH 7.3) and after pH adjustment; the solution was stored at -20 ºC and used within a month.
Extensive dimethyl suberimidate (DMS: Sigma, St Louis, MO, USA) cross-linked with the thymus core histone octamer was formed by cross-linking of histones with DMS in 2 mL NaCl and 0.1 mL sodium borate (pH 9)[18]. The cross-linked octamer was purified further by preparative gel electrophoresis and the electroelution technique. After electrophoresis, the protein complex was electroeluted for 8 h at 80–100 V at 4 ºC using dialysis tubing (3500 cut-off). The sample was then centrifuged for 15 min at 3000×g and the clear supernatant dialyzed extensively with 20 mmol/L Tris-HCl (pH 7.3) for 96 h by changing the dialysate every 6 h. After brief centrifugation, the supernatant was concentrated by a VIVASPIN filter (5000 cut-off, Vivascience, Lincoln, UK), dialyzed against 20 mmol/L Tris buffer and stored at -20 ºC until use.
The purity of the proteins was checked on 15% SDS-polyacrylamide gel electrophoresis as described by Lammeli (Figure 1B)[19]. The protein concentration was determined according to Bradford[20] using bovine serum albumin as a standard.
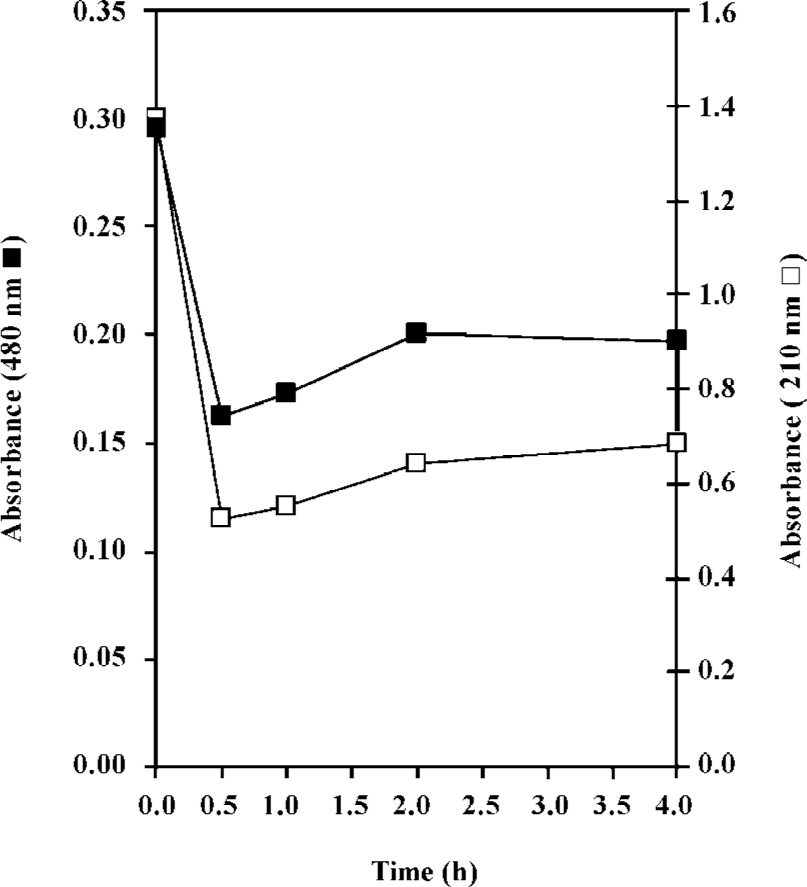
Interaction of daunomycin with histones To obtain the correct time of incubation, the proteins were incubated with various concentrations of daunomycin and after different time intervals, aliquots were taken and the absorbancies were monitored at 210 and 480 nm.
The purified core histone and CL-core were dissolved in 20 mmol/L Tris-HCl buffer (pH 7.3) and their concentration was determined spectrophotometrically. An appropriate concentration of daunomycin was incubated with the proteins (3–150 µmol/L) for 30–60 min at 23 ºC in the dark. Free daunomycin and histones were prepared in the same buffer and incubated along with the drug-histone samples under the same conditions and used as a control.
UV/Vis spectroscopy Core histone (or CL-core) and daunomycin were mixed as described above in 20 mmol/L Tris buffer (pH 7.3), and the spectrophotometric measurements were carried out at 23 ºC. The absorbancies were measured at 210, 230 and 480 nm using a UV-260 Shimadzu spectrophotometer (SHIMADZU Corporation, Kyoto, Japan) and the results were normalized for the protein concentration. Spectra were recorded between 190 and 250 nm.
Fluorescence spectroscopy The measurements were performed on a fluorescence spectrophotometer (Hitachi MPF-4, Japan) equipped with a thermostatically controlled cell holder at an ambient temperature. The monochromatic slits were set at 5 nm to reduce the intensity of the signal depending on the experiment. All samples were made in 20 mmol/L Tris-HCl (pH 7.3) at 20 ºC and a quartz fluorescence cell with a 1 cm path length was used. Protein solutions (5–10 µmol/L) were titrated with aliquots of daunomycin (0–50 µmol/L) and equilibrated until a steady emission reading was obtained. The accumulated volume of titration was less than 10 µL, so the dilution effect was negligible.
Daunomycin and the histones were prepared individually in the same buffer and used as a control. Amino acid tyrosine was also dissolved in the same buffer and its emission spectrum was recorded in the same condition and used as a control. The spectra were recorded between 290 and 370 nm after the excitation at 278 nm. The (Io–I /Io) values for each sample were normalized with respect to the fluorescence of the protein in the absence of the drug in which Io and I are fluorescence intensity before and after the addition of daunomycin, respectively.
Equilibrium dialysis The proteins in 20 mmol/L Tris-HCl buffer (pH 7.3) were dialyzed against the buffer containing serial concentrations of daunomycin using Scientific Instrument Center dialysis tubing at room temperature. The equilibrium was achieved within 72 h with the dialysis tubing. The total drug concentration (Ct) and the concentration of the free drug (Cf) in the dialysate were measured directly from the absorbance at 480 nm before and after dialysis using the extinction coefficient of 11500 mol-1· cm-1. The amount of the bound drug (Cb) was obtained from Cb=Ct-Cf. Binding parameters were determined from the plot of r/Cf versus r according to the Scatchard method[21] where r is the ratio of the bound drug to the molar concentration of proteins. The Scatchard plot gives an x-intercept of n, where n is the apparent number of binding sites and K (apparent binding constant) corresponds to the negative value of the slope of the curve. Also using this equation; 1/r=1/n+Kd/nCf , r versus Cf was drawn. The Hill coefficient (nH) was determined from the slope of the ln (Cf) versus ln (r/n–r) according to the Hill equation[22].
Results
In the present work, the binding of the anticancer drug, daunomycin, to core histones in 2 different states, core histones free in solution and core histones cross linked with DMS (CL-core), has been investigated and compared for the first time. As seen in Figure 1B, core histones represent 4 bands corresponding to histones H3, H2A, H2B and H4 and the purified protein lacks histone H1. The cross linked histone designated as CL-core, shows a histone complex with a molecular weight of about 105 kDa which is similar to the octamer size of nucleosomes. Also, the time study experiment presented in Figure 2 shows that the interaction of drug with the core histone is completed between 30 and 60 min. Therefore this incubation time range was used throughout the experiments.
Comparison of the fluorescence profiles of daunomycin interaction with core histones and CL-core The fluorescent emission spectra obtained from the interaction of daunomycin with the core and CL-core is shown in Figure 3. Figure 3A shows the fluorescence emission spectra of the core histone in the presence and absence of various concentrations of daunomycin. The fluorescence spectrum of tyrosine has also been provided for comparison. As was expected, core histones free in solution, exhibit emission spectra in the position corresponding to tyrosine with a maximum intensity at 305 nm. The addition of daunomycin to the histone solution reduces the fluorescence intensity of the protein without any red shift in the emission maxima (Imax) as the drug concentration is increased.
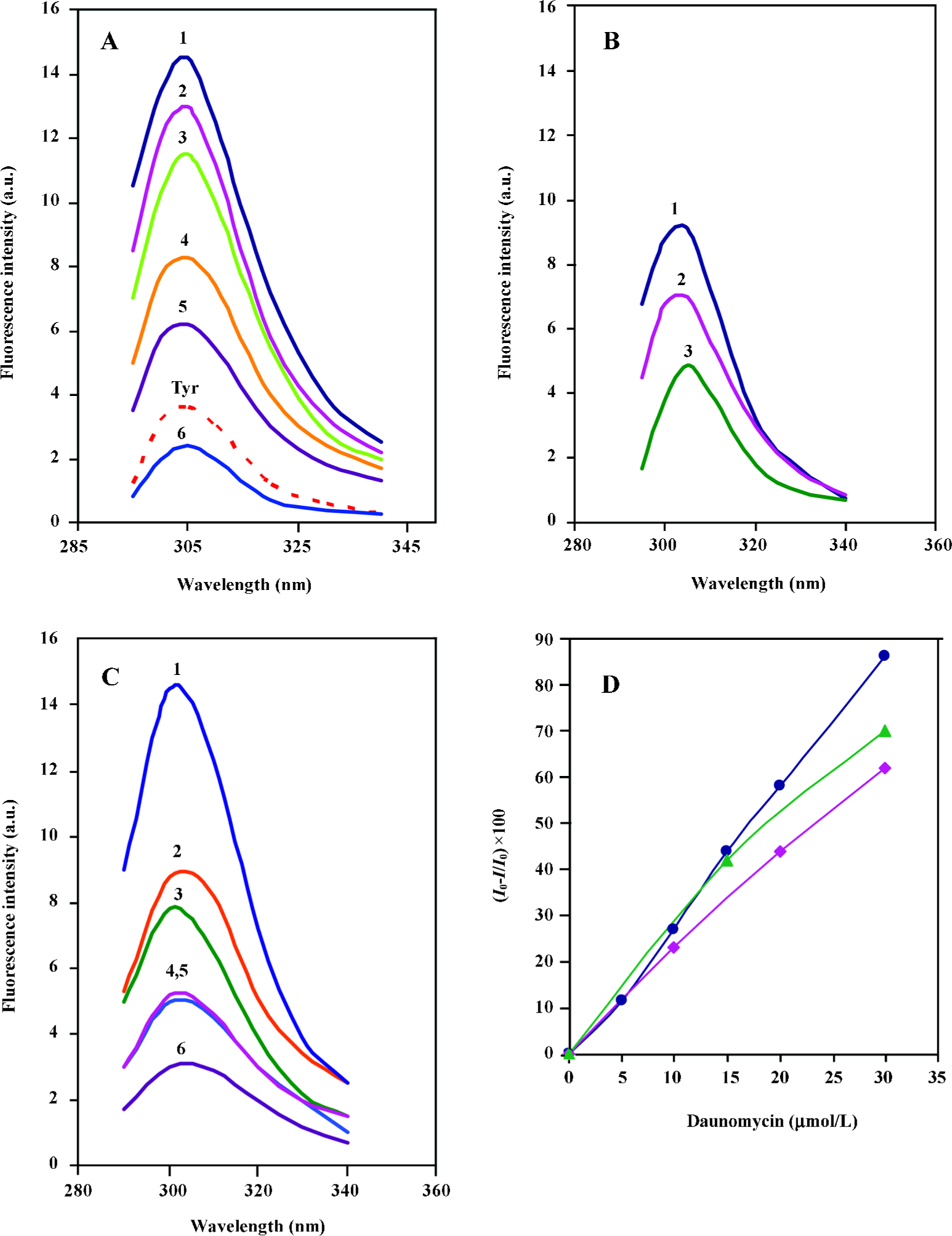
Figure 3B represents the fluorescence emission spectra of CL-core in the absence and presence of daunomycin. It is shown that the fluorescence intensity of the protein is also decreased as the drug concentration is increased, but the extent of reduction is less than that of core histones free in solution. Furthermore, for comparison, the effect of daunomycin on the fluorescence emission intensity of the individual histone pairs, H2A–H2B and H3–H4 was also investigated in the same condition. The spectra are given in Figure 3C. When the fluorescence intensities are normalized to protein concentration and Io–I/Io is calculated, the core histones still exhibit the highest emission intensity reduction and the order of intensity reduction is: core histones>H3–H4>CL-core, (Figure 3D). This clearly implies that the binding of daunomycin to core histones is dependent on their accessibility to the environment.
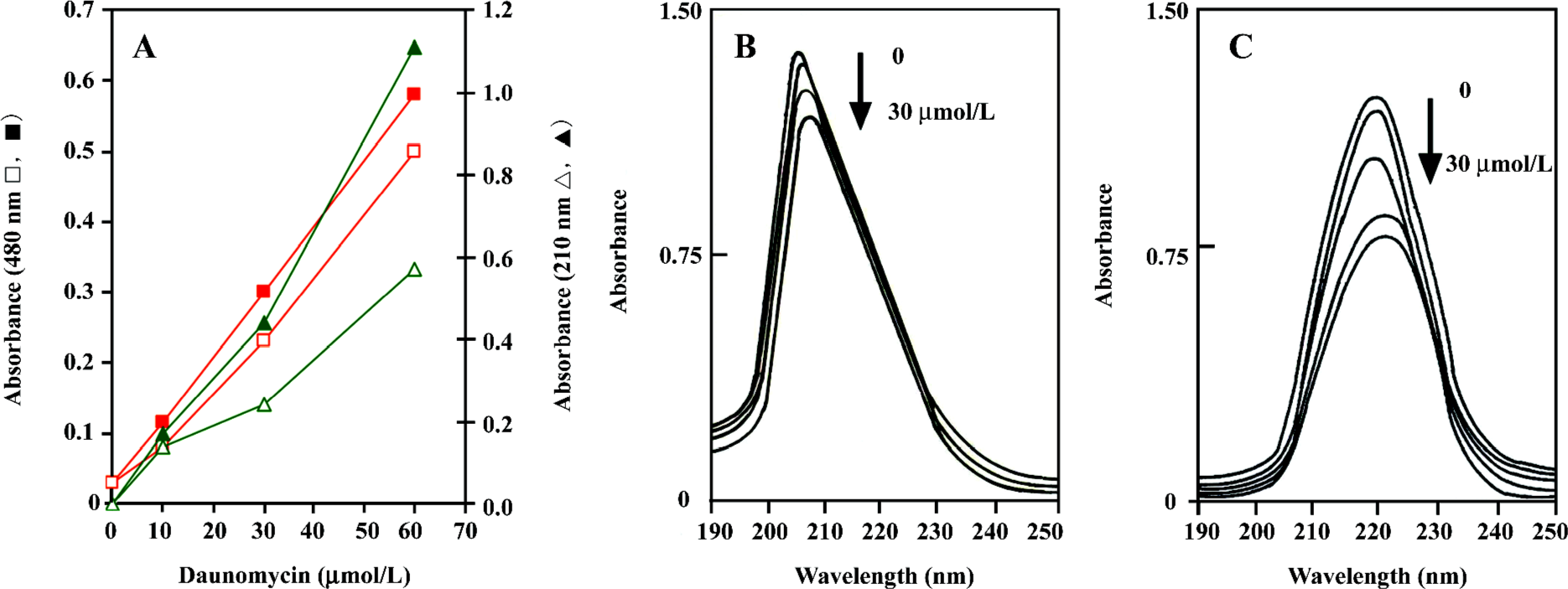
UV/Vis spectroscopy analysis Absorbance in the UV/Vis region has been successfully used for the analysis of daunomycin–DNA or linker histone[4,16] interactions. In this study, the purified proteins were incubated in the presence and absence of various concentrations of daunomycin in the dark; the changes in their absorbance at 480 and 210 nm were measured, and the data were normalized with respect to the protein concentration. Figure 4A shows the absorbance changes of the core histones as a function of the drug concentration. As can be seen, the absorbance of the core histones at 210–220 nm considerably decreased upon the addition of the drug (Figure 4A). This absorbance reduction is also observed in the pattern of 480 nm, which is exclusively related to the wavelength of daunomycin. Difference spectra patterns also confirm the above results and indicate that upon the addition of daunomycin to the protein solutions, hypochromicity occurs, while CL-core (Figure 4B) exhibits much lower hypochromicity in the presence of the drug than the core histones free in solution (Figure 4C). The results indicate that daunomycin inters the sites of histones, which are active in electron excitation.
Cooperative binding of daunomycin to core histones The binding isotherms obtained from the equilibrium dialysis is shown in Figure 5. In both cases, the Scatchard plot exhibits a cooperative binding behavior, as illustrated by the positive slope observed in the low r regions of the binding isotherms. The curve reaches a maximum at a value of r =0.8 (core histones free in solution) and r=1.3 (CL-core). A decrease in the slope is observed at higher r values. Although core and CL-core exhibit similar but not identical binding isotherms, the later shows lower r/Cf values, which is indicative of a much lower binding affinity of daunomycin to CL-core. Indeed, the binding constant (K) for the CL-core (K=5.2×105 mol-1) is relatively lower than that obtained for the core histones (K=6.5×105 mol-1). Drawing ln r/n-r against ln Cf gives a straight line with a slope of the nH (Hill coefficient). The nH values of 2.25 and 1.79 were calculated for the core histone and CL-core, respectively, which demonstrate that daunomycin has lower binding affinity to the compact structure of CL-core compared to the open structure of core histones free in solution.
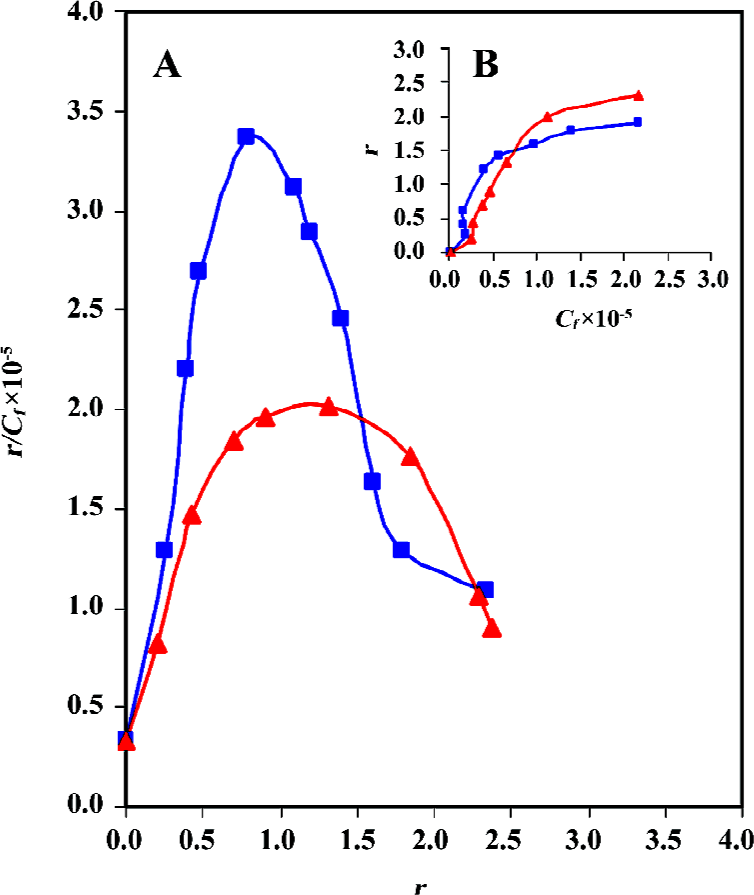
As shown the Figure 5B, a plot of r against Cf is sigmoid; that is, as Cf increases, r rises slowly at first, then rapidly and finally levels off, indicating that the system approaches to equilibrium or saturating state. Using the equation of ΔGo= -RT ln K, total macroscopic free energy values of ΔGo=-33 kcal/mol and ΔGo=-23.6 kcal/mol were obtained for the core histones and CL-core respectively. The occurrence of a negative Gibbs free energy suggests that the interaction process is exergonic.
Binding isotherms also confirm the above results. Although both samples, core histones and CL-core, show positive cooperativity, but daunomycin still shows lower binding affinity to CL-core. This is obvious when the r/Cf , K and ΔGo values are compared.
Discussion
The anticancer activity or cytotoxic effects of anthra-cycline antibiotics may involve the interaction of these drugs with the nuclear components. The binding of daunomycin to DNA and nucleosomes has been studied in detail[3–8,12]; whereas the binding of antitumor drugs to the protein components of chromatin is still questionable, and only a few reports has been published besides those dealing with the interaction of these drugs with topoisomerases[23]. During the past few years our laboratory has focused on the interaction of antitumor drug, daunomycin, with the histone H1 family (linker histones) in solution[13,14] and in chromatin[15]. The results come in support of the notion that the protein component of chromatin can also be considered as a target for the activity of these antitumor drugs. Moreover, this is the first paper demonstrating the binding behavior of anticancer drug, daunomycin, to core histones in the absence of DNA.
Structural studies have shown that core histones are comprised of 2 distinct domains. The histone fold domain (hydrophobic core), formed by 3 α-helices connected by 2 loops is involved in histone–histone and histone-DNA interactions, while the N-terminal tail domain is composed of about 15–30 highly basic amino acids, which expels out of the nucleosome surface[24,25]. Under physiological condi-tions, core histones interact with each other to form a heterotypic octamer consisting of a histone H3–H4 tetramer and two histone H2A–H2B dimers, which constitute the protein core of the basic chromatin subunit, the nuclear core particle. In solution and in the absence of DNA, the histone octamer exists in equilibrium between its constitutive H2A–H2B dimers and the H3–H4 tetramers, which have been extensively characterized[25,26].
As mentioned earlier, the fluorescence emission intensity of all proteins used here, in the absence of daunomycin exhibit a characteristic fluorescence emission intensity maximum at 305 nm corresponding to the maximum fluorescence emission of tyrosine[27]. In contrast to histone H1, which has only 1 tyrosine[28], core histones contain a higher content of tyrosine. In this case, histones H3 and H4 have 2 tyrosine and H2A and H2B contain 3–4 tyrosines (sequence analysis using Swiss Prot or Expasy programs). This aromatic amino acid is mostly located in the hydrophobic or globular part of the histones, whereas, tyrosine 38 and 122 of H2B and 39 of H2A are exposed to the solvent. Therefore the reduction in the emission intensities, in the presence of the drug, is as a result of fluorescence quenching. Most likely, in CL-core, the tyrosines are hidden inside the globular part of the protein (as a result of cross linking) providing less fluorescence quenching. Despite the ability of H2A–H2B and H3–H4 to interact with daunomycin in solution, neither of them appears to be accessible to the drug in the nucleosome environment[15]. This is confirmed by the lower affinity of daunomycin to CL-core.
Results of absorption spectroscopy have shown that daunomycin binds to both core histones and CL-core in solution and reduces the absorbancies at 210 and 480 nm (hypochromicity). Binding isotherms also confirm the above results. Although both samples, core histones and CL-core, show positive cooperativity, daunomycin still shows lower binding affinity to CL-core. This is obvious when the r/Cf , K and ΔGo values are compared. Although our results clearly demonstrate the interaction of daunomycin with the core histones in solution, but the mode of their interaction still remains obscure. From the results presented, both hydrophobic and electrostatic interactions can be speculated.
Summing up, the present study clearly demonstrates the lower affinity of daunomycin to CL-core rather than to free core histone. The result is in agreement with what we have obtained for the chromatin[12,15,29], in which the core histones covered with DNA are not accessible to the drug. We propose that, within the chromatin context, it appears that binding of daunomycin to core histones is dependent on their accessibility to the environment. This is particularly significant in the case of active chromatin and tumor cells. Neoplastic cells have a high transcription activity so that most of their chromatin exists as euchromatin. Such alteration in cancerous cells enhances the accessibility of the drug to the protein components of chromatin (core histones). This also coincides with our previous results showing that adriamycin, a similar drug to daunomycin, has higher affinity to active chromatin compared to inactive chromatin[29]. Consequently, the binding of daunomycin to DNA on one hand and to core histones on the other, produces a compact structure that inhibits chromatin metabolism, such as transcription and replication. Another explanation is that in active chromatin and in tumor cells that histones are highly acetylated, promoting the accessibility of the histones to the environment, it can be speculated that anthracycline antibiotics preferentially bind to acetylated histones, thus the complex prevents histone deacetylation. As a result, certain genes including apoptotic–related genes are continually expressed and the cells proceed into apoptosis. Although comparison of the interaction of daunomycin with acetylated and unacetylated histones, can provide important information about the real action of daunomycin at the chromatin level, extensive work needs to be elucidated.
Acknowledgements
The authors would like to thank Marzeyeh YOUSOF-MASBOOGH for her excellent research assistance.
References
- Carrion C, de Madariaga MA, Domingo JC. In vitro cytotoxic study of immunoliposomal doxorubicin targeted to human CD34 (+) leukemia cells. Life Sci 2004;75:313-28.
- Myers CE, Mimnaugh EG, Yeh GC, Sinha BK. Biochemical mechanisms of tumor cell killed by the anthracyclines, In: Lown JW, editor. Anthracenedione-based anticancer agent. Amsterdam, the Netherlands: Elsevier; 1988. p 527–69.
- Chaires JB. Molecular recognition of DNA. In: Hurley LH, Chaires JB, editors. Advances in DNA sequence-specific agent. Greenwich (CT): JAI Press; 1996. 2: 141–67.
- Barcelo F, Martorell J, Gavilanes F, Gonzalez-Ros JM. Equilibrium binding of daunomycin and adriamycin to calf thymus DNA temperature and ionic strength dependence of thermodynamic parameters. Biochem Pharmacol 1988;37:2133-8.
- Leng F, Leno GH. Daunomycin disrupts nuclear assembly and the coordinate of DNA replication in Xenopus egg extracts. J Cell Biochem 1997;64:476-91.
- Kersten W, Kersten H, Szybalski W. Physicochemical properties of complexes between DNA and antibiotics which affect ribonucleic acid synthesis. Biochemistry 1996;5:236-44.
- Fritzsohe H, Wahnert V, Chaires JB, Dattagupta N, Schlessinger FB, Crothers DM. Anthracycline antibiotics interaction with DNA and nucleosomes and inhibition of DNA synthesis. Biochemistry 1987;26:1996-2000.
- Van Holde KE. Chromatin. New York: Springer-Verlag; 1988.
- Bradbury EM. Nucleosome and chromatin structure and function. J Cell Biochem 1988; Suppl 30-31: 177–84.
- Umov FD. Chromatin as a tool for study of genome functions in cancer. Ann NY Acad Sci 2003;983:5-21.
- Chaires JB, Dattagupta N, Crothes DM. Binding of daunomycin to calf thymus nucleosomes. Biochemistry 1983;22:284-92.
- Rabbani A, Iskandar M, Ausio J. Daunomycin induced unfolding and aggregation of chromatin. J Biol Chem 1999;274:18401-6.
- Zargar SJ, Rabbani A. The effects of daunomycin antibiotic on histone H1: thermal denaturation and fluorescence spectroscopy studies. Int J Biol Macromol 2000;28:75-9.
- Zargar SJ, Rabbani A. Interaction of daunomycin antibiotic with histone H1: ultraviolet spectroscopy and equilibrium dialysis studies. Int J Biol Macromol 2002;30:113-7.
- Rabbani A, Finn RM, Thambirajah AA, Ausio J. Binding of antitumor antibiotic daunomycin to histones in chromatin and in solution. Biochemistry 2004;43:16497-504.
- Johns EW. Studies on histone: preparative methods for histone fraction from calf thymus. Biochem J 1964;92:55-9.
- Rabbani A, Goodwin GH, Jones EW. Studies on the tissue specificity of the high mobility group proteins from calf. Biochem J 1978;173:497-505.
- Thomas JO. Chemical cross linking of histones. Methods Enzymol 1989;170:549-71.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of the bacteriphage T4. Nature 1970;227:680-5.
- Bradford MM. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt Biochem 1976;72:248-54.
- Scatchard G. The attraction of proteins for small molecules and ions. Ann NY Acad Sci 1949;51:660-72.
- Ferifelder D. Physical chemistry. 2nd ed. New York: Freeman; 1982.
- Capranico G, Zunino F, Kohn KW, Pommier Y. Sequence-selective topoisomerase II inhibition by anthracycline derivatives in SV40 DNA: relationship with DNA binding affinity and cytotoxicity. Biochemistry 1990;29:562-9.
- Davey CA. Sargent Df, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol 2002;319:1097-113.
- Vaquero A, Lyol A, Reinberg D. Available from URL: , /14 / re4, 2003.http://sageke.sciencemag.org
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms,and functions. Annu Rev Biophys Biomol Struct 2002;31:361-92.
- Cowgill RW. In: Chen RF, Edelhoch H, editors, Biochemical fluorescence concepts. New York: Marcel Deker, Inc; 1976. p 441.
- Jordano J, Barbero JL, Montero F, Montero F, Franco L. Fluorescence of histones H1. A tyrosinate-like fluorescence emission in Ceratitis capitata H1 at neutral pH values. J Biol Chem 1983;258:315-20.
- Rabbani A, Jafari M. The effect of adriamycin on salt fractionated chromatin. Med J IRI 2000;13:305-10.
