Natural product juglone targets three key enzymes from Helicobacter pylori: inhibition assay with crystal structure characterization1
Introduction
Helicobacter pylori (H pylori) is a Gram-negative bacterium associated with a number of human diseases, including gastritis, peptic ulceration, and gastric cancer[1]. H pylori has been recognized as a pathogenic bacterium that chronically infects approximately 50% of the world’s human population[2]. The rapid infection of H pylori has become a severe threat against human health. Usually, the treatment of H pylori infections involves the administration of a combination of antibiotics and other drugs. However, the overuse and misuse of antibacterial agents have resulted in the generation of antibiotic resistant strains. For example, in the UK, the metronidazole resistance was found by 31.7% against H pylori isolates[3]. Accordingly, the alarming rise of antibiotic resistance among the key bacterial pathogens has strongly stimulated an urgency to develop novel antibacterial agents acting on new drug targets.
The natural product juglone (5-Hydroxy-1,4-naphthoquinone; Figure 1A), from the leaves and unripe hulls of the fruits of Juglacea, was found to demonstrate cytotoxicities when added in cell cultures[4-6]. Such an effect might result from juglone抯 involvement in the block of transcription[7], or K+ channel against cell membrane[8], the inhibition of the peptidyl-prolyl isomerase Pin1[7,9], or the generation of hydrogen peroxide[5]. In addition, juglone was found to possess antibacterial properties, including anti-H pylori and antifungal properties[10]; however, no target information has ever been disclosed.
In the present study, we reported that juglone functions as a multitargeted inhibitor against 3 key enzymes from H pylori: cystathionine γ-synthase (HpCGS), malonyl-CoA:acyl carrier protein transacylase (HpFabD), and β-hydroxyacyl-ACP dehydratase (HpFabZ). The analyzed crystal structure of the HpFabZ/juglone complex has further clarified the essential binding feature of juglone against HpFabZ at the atomic level.
It is known that different organisms display distinct spectra of transsulfuration enzymes. Most plants and microbes employ only the forward pathway from cysteine to homocysteine and methionine, and mammals carry out only the reverse transsulfuration, while fungi take up transsulfuration in both directions[11]. CGS (EC2.5.1.48), encoded by the metB gene, is a pyridoxal 5′-phosphate-dependent enzyme responsible for the γ-replacement reaction of an activated form of L-homoserine with L-cysteine, leading to L-cystathionine. For microorganisms, such a reaction is the first step involved in the transsulfuration pathway that converts L-cysteine into L-homocysteine. Since CGS is absent in non-ruminant animals that require a dietary source of L-homocysteine or L-methionine[12,13], it has thus been regarded as an attractive target for antibiotic discovery[14]. However, the metB gene seems to be unessential for H pylori growth as reported by Salama et al[15].
Fatty acid biosynthesis (FAS) is an essential pathway for the survival of the organism since fatty acid is the major component of cell membranes and possesses important biological functions. In nature, according to the enzymes involved in the pathway, fatty acid biosynthesis is divided into 2 types: type I (FAS I) and type II (FAS II)[16-18]. In the FAS I system, which found in animals, the biosynthesis of fatty acid is catalyzed by a multi-enzyme, which is a single polypeptide with 8 distinct domains. However, in the FAS II system, the reactions are carried out by a series of structurally dissociated enzymes as discovered in most bacteria[16,17]. Due to the large differences between these 2 FAS systems, enzymes like FabD (EC2.3.1.39) and FabZ (EC4.2.1.60) involved in type II fatty acid biosynthesis have been developed as potential targets for the discovery of antibacterial agents[17]. FabD catalyzes the transfer of a malonyl moiety from malonyl-CoA to holo-ACP, forming malonyl-ACP as the elongation substrate for the fatty acid biosynthesis[19-22], while FabZ is the primary dehydratase that participates in the elongation cycles of saturated and unsaturated fatty acid synthesis[23-26].
It is expected that our current work might help understand the possible antibacterial mechanism for juglone and provide useful structural information for the discovery of an anti-H pylori agent by using juglone as a potential multitargeted lead compound.
Materials and methods
Materials H pylori strain SS1 was maintained at our institute. The Escherichia coli host strain BL21(DE3) was purchased from Stratagene (Germany). The natural product juglone was from the in-house chemical library established in our laboratory. All other chemicals used were of reagent grade or ultra-pure quality.
Expression and purification of HpCGS The HpCGS enzyme was cloned, expressed, and purified according to our recently published work[27]. Briefly, HpmetB was PCR-amplified from H pylori SS1 strain genomic DNA and ligated into a pET28b expression vector (Novagen, Germany) and transformed into BL21(DE3) after restriction digestion. The correct clones were expressed at 37 °C, and the purified HpCGS enzyme was obtained by using buffer C (20 mmol/L Tris-HCl, pH 8.0, 500 mmol/L NaCl, and 120 mmol/L imidazole) as elution buffer.
Expression and purification of HpFabD The cloning, expression, and purification of HpFabD were performed according to our published description[28]. The purified HpFabD protein was dialyzed against buffer A (20 mmol/L Tris-HCl, pH 8.0) to remove imidazole.
Expression and purification of HpFabZ The cloning, expression, and purification of HpFabZ were based on our recently published work[29]. The purified protein was obtained by dialysis against buffer B (20 mmol/L Tris-HCl, pH 8.0, 500 mmol/L NaCl, and 1 mmol/L EDTA).
HpCGS inhibition assay The HpCGS enzyme inhibition assay was carried out according to our recently published study[27]. The IC50 value of juglone against HpCGS was obtained by fitting the data to a sigmoid dose-response equation using Origin software (OriginLab, Northampton, Massachusetts, USA). Inhibitor type and inhibition constants Ki and Ki were determined by the double-reciprocal (Lineweaver-Burk) plot, and the Vmax plot and slopes of the lines from the double-reciprocal plot were used as a function of inhibitor concentrations.
HpFabD inhibition assay The HpFabD enzyme inhibition assay was performed based on our reported approach[28]. To determine the IC50 value of juglone against malonyl-CoA, HpFabD was incubated for 5 min at room temperature with different concentrations of juglone (0?100 μmol/L) in a total volume of 100 μL. To investigate the inhibition mode of juglone against malonyl-CoA, different concentrations of juglone (0, 8, 10 μmol/L) were used, and the reaction was initiated by the addition of malonyl-CoA (5?30 μmol/L). The Ki value was obtained by the Dixon plot.
HpFabZ inhibition assay The enzymatic inhibition assay of the HpFabZ enzyme was monitored by using the reported spectrophotometric method[26,29,30]. The IC50 value of the inhibitor was estimated by fitting the inhibition data to a dose-dependent curve using a logistic derivative equation[31]. The inhibitor mechanism was determined in the presence of various inhibitor concentrations (0‒50 μmol/L). After 2 h incubation, the reaction was started by the addition of crotonoyl-CoA (10‒250 μmol/L). The Ki value was obtained from the Dixon plot and secondary plots.
Crystallization and data collection The crystallization of the HpFabZ/juglone complex was carried out according to our previous work[30]. The purified HpFabZ enzyme in 20 mmol/L Tris-HCl (pH 9.0) and 500 mmol/L NaCl was concentrated to ~10 μg/mL. For crystallization, 1 μL of the enzyme was mixed with an equal volume of the reservoir solution containing 2 mol/L sodium formate and 0.1 mol/L sodium acetate trihydrate at pH 3.6‒5.6, and benzamidine-HCl was added to a final concentration of 2% (w/v). The mixture was equilibrated against 500 μL of the reservoir solution at 277K by the hanging-drop vapor-diffusion method. Crystals of dimensions 0.5×0.3×0.3 mm3 were obtained after 7 d. Juglone was added to the original drop to a final concentration of ~20 mmol/L, and the crystals were soaked for 24 h.
Diffraction data were collected at 100K using CuKα X-ray with a Rigaku R-AXIS IV++ image plate (Rigaku Corp, Tokyo, Japan). Before the crystals were flash-frozen in liquid nitrogen, the drop was dehydrated against 500 μL reservoir solution containing 4 mol/L sodium formate for 24 h. The data were processed using HKL2000[32]. The crystallographic statistics are summarized in Table 1. The structure was solved by the molecular replacement approach using the crystal structure of HpFabZ as the search model and refined by the program CNS[33]. Electron density interpretation and model building were performed using the computer graphics program “COOT”[34].

Full table
Results
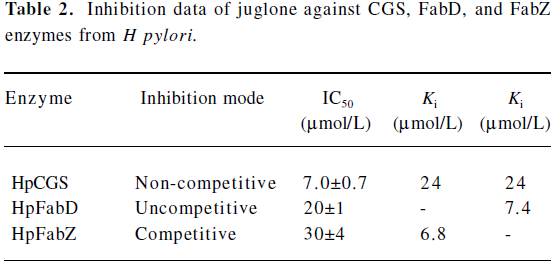
Juglone is a multitargeted inhibitor against CGS, FabD, and FabZ enzymes Inhibition against CGS enzyme as evaluated from the enzymatic assay, the natural product juglone was identified to show strong inhibitory activity against HpCGS with an IC50 of 7.0?0.7 μmol/L (Table 2). In a separate control experiment by directly using 2 mmol/L α-ketobutyrate as the substrate, juglone showed no inhibition activity against D-2-hydroxyisocaproate dehydrogenase (HO-HxoDH) at its concentration, even up to 50 μmol/L, further confirming that juglone is a HpCGS inhibitor. The Lineweaver-Burk plot-based analysis indicated that juglone prevented the binding of O-succinyl-L-homoserine (L-OSHS) to HpCGS in a non-competitive fashion (Figure 2A), and the inhibition constants Ki of 24 μmol/L and Ki of 24 μmol/L (Table 2) were determined from the Dixon plot and secondary plots.
Full table
Inhibition against HpFabD As indicated in Table 2, juglone could inhibit HpFabD with an IC50 against malonyl-CoA at 20?1 μmol/L as a non-competitive inhibitor (Figure 2B). The αKi value of 7.4 μmol/L (Table 2) was thereby obtained by the Dixon plot.

Inhibition against HpFabZ As indicated in Figure 2C, besides targeting CGS and FabD, juglone was also detected as a competitive inhibitor against HpFabZ by an IC50 of 30? 4 μmol/L (Table 2) with respect to the substrate crotonoyl-the apparent value of Km at different inhibitor concentrations as a function of the inhibitor concentration.
Crystal structure analysis of the HpFabZ/juglone complex In order to gain the essential inhibition mechanism at the atomic level for juglone against these 3 enzymes, we tried crystallizing the enzymes in complex with juglone and succeeded in obtaining the crystal of the HpFabZ/juglone complex, which was well determined against 2.4 A level data (PDB code: 3B7J, Figure 3 and Table 1). As shown in the analyzed complex structure, juglone binds to HpFabZ in 2 binding models similar to our previous binding model for HpFabZ binding to the inhibitor[30] (Figure 4). In model A, juglone fits into the groove around the entrance (Figure 4A) and locates between the phenol ring of Tyr100 and the pyrrolidine ring of Pro112', forming a sandwich structure (the prime indicates a residue from the other subunit in the dimer). As shown in Figure 4B, the carbonyl oxygen of juglone forms H-bonds with the nearby water chain, which also forms H-bonds with the back bone carbonyl oxygen atoms of Phe65', Ile111', Val 146', and Phe101, as well as the back bone nitrogen of Val113'. In model B, juglone entered into the middle of the tunnel near the active site of HpFabZ (His58 and Glu72'; Figure 4C) and was stabilized via the hydrophobic interactions between residues Leu21, His23, Ala75, Phe83, Ile98, Val 99, and Phe59'. At this stage, the carbonyl oxygen of juglone could form H-bonds with the nearby water chain that also forms H-bonds with the back bone carbonyl oxygens of Ala75, the Nε2 of His 23 and His58', and Oε1 and Oε2 of Glu72 (Figure 4D).


Discussion
Recently, the rapid infection of H pylori has become a severe threat to human health, and the discovery of new, effective drugs has attracted more and more attention. Many antibacterial agents used for standard pathogens in clinics are subject to high-level resistance resulting from single-step mutations from target enzymes, whereas multitargeted agents might display low potential for rapid endogenous resistance development in pathogenic bacteria[35].
It is reported that juglone could strongly inhibit H pylori growth at a low MIC of 1.6 μg/mL[36,37], although no acting target information has ever been disclosed. In the current work, we discovered that juglone functions as a multitargeted inhibitor against 3 key enzymes, HpCGS, HpFabD, and HpFabZ, which provides some clues for the inhibitory mechanism underlying juglone’s anti-H pylori activity.
In the last 10 years, many CGS and FabZ inhibitors have been discovered. However, most of the inhibitors are single-targeted, except some flavonoid derivatives (luteolin and (-)-catechin gallate), which were reported to inhibit Plasmodium falciparum by acting as multitargeted inhibitors against FabG, FabZ, and FabI of Plasmodium falciparum[38].
In our previous work[27], we reported that the natural products α-lapachone and 9-hydroxy-α-lapachone demonstrated inhibitory activities against HpCGS. Considering the structure similarity between these 2 compounds and juglone, each of them contains a common quinone group (Figure 1). Although the natural products α-lapachone and 9-hydroxy-α-lapachone exhibited no inhibition activities against HpFabD and HpFabZ, as evaluated by the inhibition assay, the quinone group seemed to have some links to the CGS enzyme inhibition.
The natural product corytuberine was our first published HpFabD inhibitor[28]. Similar to corytuberine, juglone functions in an uncompetitive mode to inhibit HpFabD (Table 2). In addition, the inhibition study indicated that juglone was a competitive inhibitor of HpFabZ, suggesting that juglone may interfere with the binding of substrate crotonoyl-CoA, which could further be proved by the inhibitor binding manner in the complex structure. The binding affinity of juglone to HpFabZ (Ki=6.8 μmol/L) was higher than that to HpCGS or HpFabD, which might be attributed to the competitive inhibition type of juglone against HpFabZ.
Previously[30], we reported 2 crystal structures of HpFabZ in complex with 2 small molecular inhibitors, and 2 binding models of an inhibitor against HpFabZ were supposed. In binding model A, the inhibitor is sandwiched between the phenol ring of Tyr100 and the pyrrolidine ring of Pro112', and forms a π−π interaction. In binding model B, the inhibitor locates into the middle of the tunnel via hydrophobic interactions. Juglone also fits in these 2 models (Figure 3), of which the position in model A is corresponding to the groove around the entrance, and the position in model B is corresponding to the active site of the tunnel near the catalytic residues His58 and Glu72, in agreement with the competitive inhibitory properties of juglone against HpFabZ determined by the inhibition type study. However, in comparison with this previous HpFabZ-inhibitor crystal structure analysis, 2 major differences are found for juglone binding to HpFabZ. First, in the binding model of HpFabZ/juglone, besides the π−π or hydrophobic interactions, juglone was also stabilized by the H-bonds formed by the water chain between the enzyme and juglone. The other major difference is that in binding model B, juglone is inserted deeper to the tunnel (near Phe83) with a more suitable hydrophobic environment for the binding, different from the previously reported HpFabZ-inhibitor binding case, where the inhibitor formed a sandwich-like conformation with Ile98 and His59' of HpFabZ. It is thus espected that the information based on the HpFabZ/juglone complex would be useful for structure-guided drug design which might provide novel competent HpFabZ inhibitors.
In summary, in the present study, we have discovered that the natural product juglone is a multitargeted inhibitor against 3 key enzymes CGS, FabD, and FabZ from H pylori. The obtained crystal structural data for the HpFabZ/juglone complex has further clarified the essential binding feature of juglone against HpFabZ at the atomic level. Although further experiments, such as a cell labeling assay[39], are needed to determine whether or not these 3 enzymes are targets responsible for juglone’s anti-H pylori activity, juglone could hopefully serve as a potent lead compound for further inhibitory development as stated, and that the multi-targeted inhibitors might provide low potential for rapid endogenous resistance development[35].
References
- Dubreuil JD, Giudice GD, Rappuoli R. Helicobacter pylori interactions with host serum and extracellular matrix proteins: potential role in the infectious process. Microbiol Mol Biol Rev 2002;66:617-29.
- Cover TL, Blaser MJ. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med 1996;41:85-117.
- Cameron EA, Powell KU, Baldwin L, Jones P, Bell GD, Williams SG. Helicobacter pylori: antibiotic resistance and eradication rates in Suffolk, UK, 1991 2001. J Med Microbiol 2004;53:535-8.
- Paulsen MT, Ljungman M. The natural toxin juglone causes degradation of p53 and induces rapid H2AX phosphorylation and cell death in human fibroblasts. Toxicol Appl Pharmacol 2005;209:1-9.
- Inbaraj JJ, Chignell CF. Cytotoxic action of juglone and plumbagin: a mechanistic study using HaCaT keratinocytes. Chem Res Toxicol 2004;17:55-62.
- Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, et al. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ 2000;11:409-16.
- Chao SH, Greenleaf AL, Price DH. Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res 2001;29:767-73.
- Varga Z, Bene L, Pieri C, Damjanovich S, Gaspar R. The effect of juglone on the membrane potential and whole-cell K+ currents of human lymphocytes. Biochem Biophys Res Commun 1996;218:828-32.
- Hennig L, Christner C, Kipping M, Schelbert B, Rucknagel KP, Grabley S, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry 1998;37:5953-60.
- Alice MC, Tannis MJ, Charles DH. Antimicrobial activity of juglone. Phytotherapy Research 1990;4:11-4.
- Clausen T, Huber R, Prade L, Wahl MC, Messerschmidt A. Crystal structure of Escherichia coli cystathionine gamma-synthase at 1.5 A resolution. EMBO J 1998;17:6827-38.
- Soda K. Microbial sulfur amino acids: an overview. Methods Enzymol 1987; 143: 453 9.
- Aitken SM, Kim DH, Kirsch JF. Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for the elimination and substitution reactions. Biochemistry 2003;42:11297-306.
- Wahl MC, Huber R, Prade L, Marinkovic S, Messerschmidt A, Clausen T. Cloning, purification, crystallization, and preliminary X-ray diffraction analysis of cystathionine gamma-synthase from E coli. FEBS Lett 1997;414:492-6.
- Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol 2004;186:7926-35.
- Campbell JW, Cronan JE. Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu Rev Microbiol 2001;55:305-32.
- White SW, Zheng J, Zhang YM, Rock CO. The structure biology of type II fatty acid biosynthesis. Annu Rev Biochemistry 2005;74:791-831.
- Magnuson K, Jackowski S, Rock CO, Cronan JE. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev 1993;57:522-42.
- Williamson IP, Wakil SJ. Studies on the mechanism of fatty acid synthesis. XVII. Preparation and general properties of acetyl coenzyme A and malonyl coenzyme A-acyl carrier protein transacylases. J Biol Chem 1966;241:2326-32.
- Ruch FE, Vagelos PR. The isolation and general properties of Escherichia coli malonyl coenzyme A-acyl carrier protein transacylase. J Biol Chem 1973;248:8086-94.
- Verwoert II, Verbree EC, van der Linden KH, Nijkamp HJ, Stuitje AR. Cloning, nucleotide sequence, and expression of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase. J Bacteriol 1992;174:2851-7.
- Kutchma AJ, Hoang TT, Schweizer HP. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J Bacteriol 1999;181:5498-504.
- Mohan S, Kelly TM, Eveland SS, Raetz CR, Anderson MS. An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J Biol Chem 1994; 269: 32 896-903.
- Heath RJ, Rock CO. Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J Biol Chem 1996; 271: 27 795-801.
- Pillai S, Rajagopal C, Kapoor M, Kumar G, Gupta A, Surolia N. Functional characterization of beta-ketoacyl-ACP reductase (FabG) from Plasmodium falciparum. Biochem Biophys Res Commun 2003;303:387-92.
- Sharma SK, Kapoor M, Ramya TN, Kumar S, Kumar G, Modak R, . Identification, characterization, and inhibition of Plasmodium falciparum beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J Biol Chem 2003; 278: 45 661-71.
- Kong YH, Wu DL, Bai HY, Han C, Chen J, Chen LL, et al. Enzymatic characterization and inhibitor discovery of a new cystathionine γ-synthase (CGS) from Helicobacter pylori. J Biochem (Tokyo) 2008;143:59-68.
- Liu WZ, Han C, Hu LH, Chen KX, Shen X, Jiang HL. Characterization and inhibitor discovery of one novel malonyl-CoA: acyl carrier protein transacylase (MCAT) from Helicobacter pylori. FEBS Lett 2006;580:697-702.
- Liu WZ, Luo C, Han C, Peng SY, Yang Y, Yue J, et al. A new beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Helicobacter pylori: molecular cloning, enzymatic characterization, and structural modeling. Biochem Biophys Res Commun 2005;333:1078-86.
- Zhang L, Liu WZ, Hu TC, Du L, Luo C, Chen KX, et al. Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J Biol Chem 2008;283:5370-9.
- Chen LL, Gui CS, Luo XM, Yang QG, Gunther S, Scandella E, et al. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J Virol 2005;79:7095-103.
- Otwinowski Z, Minor W. Methods in Enzymology 1997;276:307-26.
- Brunger AT, Adams PD, Clore GM, DeLano W L, Gros P. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998;54:905-21.
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126-32.
- Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Discov 2007;6:41-55.
- Inatsu S, Ohsaki A, Nagata K. Idebenone acts against growth of Helicobacter pylori by inhibiting its respiration. Antimicrob Agents Chemother 2006;50:2237-9.
- Park BS, Lee HK, Lee SE, Piao XL, Takeoka GR, Wong RY, et al. Antibacterial activity of Tabebuia impetiginosa Martius ex DC (Taheebo) against Helicobacter pylori. J Ethnopharmacol 2006;105:255-62.
- Tasdemir D, Lack G, Brun R, Ruedi P, Scapozza L, Perozzo R. Inhibition of Plasmodium falciparum fatty acid biosynthesis: evaluation of FabG, FabZ, and FabI as drug targets for flavonoids. J Med Chem 2006;49:3345-53.
- Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 2006;441:358-61.
- Delanoue WL. The PyMOL Molecular Graphics System. San Carlos, CA: DelanoScientific, 2000.
