Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients
Introduction
During post-synthetic DNA modification, the methylation of DNA and the enzymes regulating methylation are crucial for the regulation of gene expression, and may contribute to the understanding of the pathogenesis of autoimmune diseases[1–3]. DNA methyltransferase (DNMT), particularly for DNMT1, is responsible for the methylation of DNA molecules and converts cytosine to methylcytosine (mC)[4,5]. mC in the CpG islands in the regulatory regions of a gene, such as the promoter and the first exon of DNA sequences, usually result in transcriptional silences, whereas their hypomethylation often leads to the overtranscription of the gene[6]. These epigenetic alterations in genomic DNA have been implicated in the pathogenic process of autoimmune diseases, such as systemic lupus erythematosus (SLE)[7,8].
SLE is characterized by the overproduction of autoantibodies, which are attributed to the abnormal hyperactivity of T and B cells[9,10]. Interleukin (IL)-4 is a growth and differential factor, contributing to the development of Th2 responses. IL-4 promotes B-cell differentiation and enhances antibody production. IL-6 stimulates B-cell proliferation, differentiation, and antibody production. Importantly, high levels of IL-6 mRNA and protein expressions were detected in freshly isolated lymphocytes from SLE patients, and high levels of serum IL-6 are correlated with disease activity and severity. However, little is known about the mechanism(s) underlying the expression of these cytokines in SLE patients.
Notably, lower activity of DNMT1 and lower levels of DNMT1 expression were found in T cells from patients with SLE, which is associated with the DNA hypomethylation of CD4+ T cells from SLE patients[11]. Furthermore, the treatment of T cells from healthy individuals with SLE-inducer, 5-azacytidine (5-azaC), and other DNA methylation inhibitors, has been found to induce the demethylation of mC, leading to autoreactivity[12,13]. Importantly, the hypomethylation of specific genes in T cells from SLE patients has been demonstrated to upregulate the expression of lymphocyte function-associated antigen-1 (LFA-1), CD70, and perforin[14–16]. Hence, DNA methylation and chromatin structure, as well as other SLE inducers, can regulate the expression of SLE-related genes. Given that IL-4 is an important regulator in the development of SLE and that high levels of IL-6 contribute to the pathogenesis of SLE, we hypothesized that the hypomethylation of IL-4 and -6 genes, particularly in the promoter regions in T cells, could upregulate the expressions of IL-4 and -6 in SLE patients.
To test this hypothesis, we purified T cells from SLE patients and healthy volunteers, activated them in vitro, and characterized the methylation levels in the IL-4 and -6 promoters. We found that the mC/G contents were significantly reduced in T cells from SLE patients, as compared with that from the healthy volunteers, which was inversely correlated to the levels of IL-4 and -6 transcripts, as well as the severity of SLE. Furthermore, we found that the treatment of healthy T cells with 5-azaC induced demethylation of the CpG islands in the IL-4 and -6 promoter regions, similar to T cells spontaneously developed in SLE patients. We discussed the implication of these findings in the pathogenesis of SLE.
Materials and methods
Patients Twenty SLE patients and 10 healthy volunteers were recruited from the outpatient clinics and inpatients services at the Second Affiliated Hospital of Sun Yat-sen University (Guangzhou, China). Patients with SLE met the criteria of the American College of Rheumatology and were scored by the SLE disease activity index (SLEDAI). Their clinical characteristics are shown in Table 1. Written informed consent was obtained from individual patients, and the experimental protocols were approved by the Institutional Research Board of Sun Yat-sen University.

Full table
T-cell purification and cell culture Peripheral mononuclear cells were isolated from the blood by density gradient centrifugation. Their T cells were purified by a nylon fiber column. The T cells with purity greater than 90%, as determined by Facial Action Coding System (FACS) analysis with fluorescein isothiocyanate-conjugated anti-CD3, were used for the following experiments. T cells from individual patients at 1×106 cells/mL were stimulated with 1 µg/mL phytohemagglutinin (PHA; Sigma, St Louis, MO, USA) in 10% fetal calf serum RPMI-1640 for 24 h and then expanded with 50 U/mL IL-2 (Hyclone, Logan, UT, USA) for an additional 72 h. Following activation with PHA, partial T cells from the healthy controls were expanded with IL-2 in the presence or absence of 1 µmol/mL 5-azaC (Sigma, USA) to determine whether 5-azaC treatment could induce demethylation in the IL-4 and -6 promoter regions of the T cells and change the expression of these cytokines. After expansion, these T cells were harvested for the following experiments.
Semiquantitative RT-PCR Total cellular RNA from 5×106 activated T cells was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) and reversely transcripted into cDNA. The levels of IL-4, and -6 mRNA transcripts were determined by semiquantitative RT–PCR. The specific primers used for the amplification of IL-4 were: forward, 5'-TGAAC AGCCT CACAG AGCAG-3' and reverse, 5'-GCGAG TGTCC TTCTC ATGGT-3'; IL-6: forward, 5'-ATGCA ATAAC CACCC CTGAC-3' and reverse, 5'-TAAGT TCTGT GCCCA GTGGA-3'; and β-actin: forward, 5'-GATGA GATTG GCATG GCTTT-3' and reverse, 5'-CTCAA GTTGG GGGAC AAAAA-3'. The cDNA templates were first denatured at 94 °C for 5 min and the PCR reactions were subjected to 35 cycles at 94 °C for 1 min, 55 °C for 0.5 min, and 72 °C for 1 min followed by extension at 72 °C for 10 min. After PCR, an equal amount of the PCR products were separated by gel electrophoresis. The DNA fragments generated by PCR were visualized under UV light and photographed with the UVIgelstartMw System (UVITec, CAMBRIDGE, UK).The levels of IL-4 and -6 mRNA transcription were determined by densitometry scanning and expressed as the levels relative to β-actin mRNA transcription.
Bisulfate modification of genomic DNA Genomic DNA was extracted from 5×106 T cells and treated with sodium bisulfate according to the protocol of the CpGenome fast DNA modification kit (Chemicon, Temecula, CA, USA). In brief, 1.0 µg purified DNA was denatured with 7.0 µL 3 mol/L NaOH and 100 µL water at 37 °C for 10 min. The DNA was modified with 550 µL DNA modification reagent, which was freshly prepared at 50 °C overnight. The DNA modification reagent containing sodium salt of bisulfate ion makes the unmethylated cytosine sulfonated and hydrolytically deaminated, yielding a uracil sulfonate intermediate. The DNA was bound to a column to remove free salts. The conversion to uracil was completed by alkaline desulfonation and desalting with 70% EtOH. The DNA was finally eluted from the column using elution buffer. The DNA was either used immediately as the templates for PCR or stored at –20 °C.
Nested PCR and direct sequencing The DNA fragments of the IL-4 or -6 promoter region were amplified by nested PCR. The specific primers used for the IL-4 promoter were: first forward, 5'-CAGTG CTGGG GTAGG A-3' and reverse, 5'-ACACC ATAAT TTGCT CTTTA-3'; second forward, 5'-TTTCC ATGAC AGGAC AGT-3' and reverse, 5'- ACACC ATAAT TTGCT CTTT-3'; IL-6 promoter: first forward, 5'-AAGTA AAGGA AGAGT GGTTC-3' and reverse, 5'-CACCC CTCCC TCACA-3'; and second forward, 5'-TAAAG GAAGA GTGGT TCTGC-3' and reverse, 5'-GGATT TCCTG CACTT ACTTG-3'. The PCR reactions were performed by 1 cycle at 94 °C for 6 min, 55 °C for 1 min, and 72 °C for 1 min, and then subjected to 38 cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min, followed by 1 final cycle at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 11 min. The yielded DNA fragments of the IL-4 or -6 promoter in T cells from individual patients were purified and sequenced directly. The contents of unmethylated cytosine in the CpG islands of the IL-4 or -6 promoter were quantitatively determined and expressed as the percentage of unmethylated cytosine.
Statistical analysis The results are presented as mean±SEM. Group comparisons with quantitative data were statistically analyzed using Student’s t-test, while qualitative data were calculated using the χ2-test. The association between the SLEDAI and levels of hypomethylation in the IL-4 or -6 promoter of T cells from SLE patients was analyzed by logistic regression. All statistical analyses were performed using SPSS software for Windows (SPSS, Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
Higher levels of IL-4 and -6 mRNA transcripts in lupus T cells IL-4 and -6 are crucial cytokines for the regulation of SLE pathogenesis. However, little is known about the relative levels of these cytokines expressed by activated peripheral T cells in Chinese SLE patients. To address this issue, peripheral T cells were isolated from groups of SLE patients and healthy volunteers. After activation in vitro, the levels of IL-4 and -6 mRNA transcripts were characterized by RT–PCR (Figure 1). The levels of IL-4 mRNA transcripts in T cells from Chinese SLE patients were 2-fold higher than that in the healthy controls (1.30±0.56 vs 0.46±0.14, P<0.01). A similar pattern of IL-6 mRNA transcripts was observed in these 2 groups of patients (1.23±0.464 vs 0.59±0.1, P<0.01). These data suggest that higher levels of IL-4 and -6 are expressed by activated T cells in Chinese SLE patients.

Treatment of healthy T cells with 5-azaC enhances IL-4 and -6 mRNA transcriptions Treatment with 5-azaC has been shown to induce DNA demethylation and upregulate gene expression[17–19]. Next, we tested whether the treatment of activated T cells with 5-azaC could modulate the IL-4 and -6 mRNA transcription. Peripheral T cells from healthy patients were activated with PHA in vitro and then expanded with IL-2 in the presence or absence of 5-azaC for 72 h. Subsequently, the levels of IL-4 and -6 mRNA transcripts were determined by RT-PCR (Figure 2). In the absence of 5-azaC treatment, moderate levels of IL-4 and -6 transcripts were detected. In contrast, significantly higher levels of IL-4 (P=0.013) and IL-6 (P=0.001) transcripts were observed in T cells treated with 5-azaC. Therefore, the treatment of healthy activated T cells with 5-azaC enhanced the IL-4 and -6 mRNA transcriptions, which may be associated with the demethylation of the IL-4 and -6 genes.

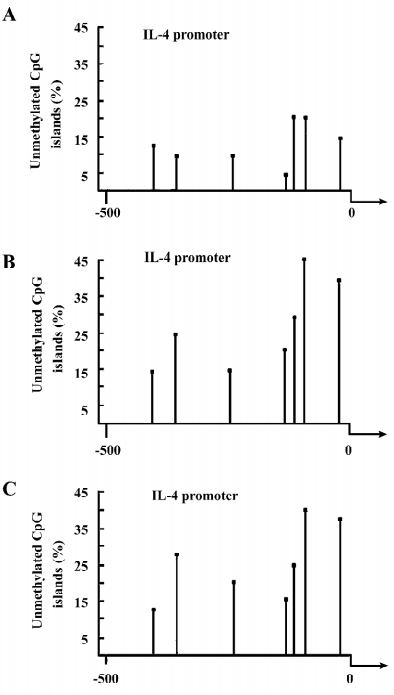
Treatment of healthy T cells with 5-azaC induces the demethylation of mC in the IL-4 and -6 promoters Given that demethylation of mC in the regulatory region of the gene, such as the promoter, usually promotes high levels of gene expression, we further examined whether the treatment of activated T cells with 5-azaC could induce the demethylation of mC in the IL-4 and -6 promoters. Activated T cells (originally from healthy volunteers) were treated with or without 5-azaC for 3 d. Their genomic DNA were extracted and used as templates for the characterization of the methylation statues of the IL-4 or -6 promoter by bisulfate modification and nested PCR. There are 7 or 10 CpG pairs potentially methylated and located at positions –419, –367, –229, –101, –92, –79, and –56 in the IL-4 promoter or –917, –824, –803, –787, –701, –596, –468, –351, –263, and –115 in the IL-6 promoter, respectively, near to the transcription start site of the genes. As shown in the Figure 3A and 3B, the percentage of the CpG demethylation in the IL-4 promoter in the 5-azaC-treated T cells was higher than that of the control T cells. Interestingly, while approximately 5% of the CpG was unmethylated at position –101 in the IL-4 promoter in the untreated control T cells, nearly 22% of the CpG islands at the same position were demethylated in the 5-azaC-treated T cells. The average unmethylation of the IL-4 promoter in the 5-azaC-treated T cells was significantly higher than that of the untreated control T cells (0.28±0.09 vs 0.09±0.04, P<0.05; Figure 4A). Similarly, the treatment of healthy T cells with 5-azaC induced higher levels of demethylation of the CpG islands in the IL-6 promoter (Figure 5A,5B) with an average percentage of unmethylated CpG (0.29±0.11 vs 0.12±0.05; Figure 4B, P<0.05). Therefore, treatment with 5-azaC to inhibit DNA methyltransferase activity induced significantly increased levels of demethylation in the CpG islands of the IL-4 and -6 promoters, which may contribute to higher levels of IL-4 and -6 mRNA transcriptions in T cells.


Alternatively, in order to further exclude the influence of PHA, human T cells were cultured in the presence or the absence of PHA for 72 h. IL-4 and -6 expressions and their promoter methylation in unstimulated T cells were compared with PHA-stimulated T cells. PHA did not cause significant changes in the expression and promoter methylation status of IL-4 and -6, suggesting that PHA does not account for the differences in expression and promoter methylation status.
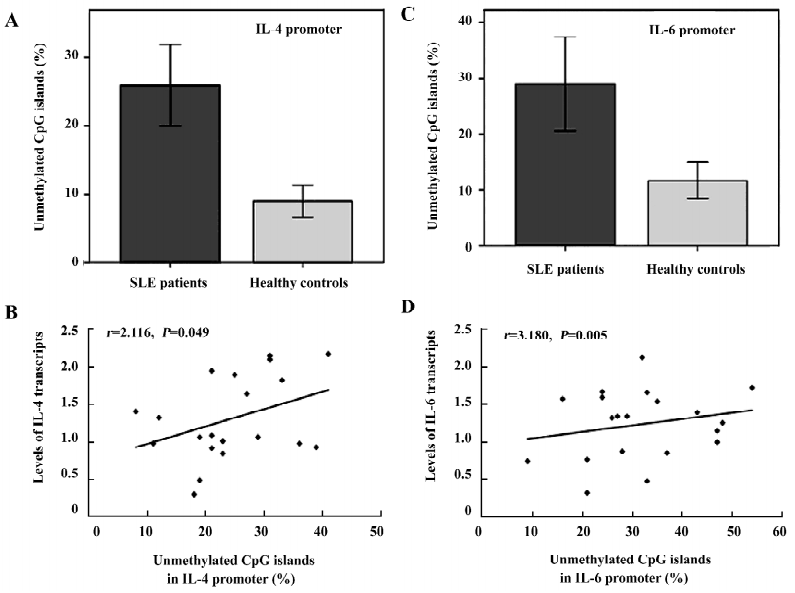
Hypomethylation of the IL-4 and -6 promoters in lupus T cells Higher levels of IL-4 and -6 transcripts were detected in lupus T cells, which were possibly mediated by the hypomethylation of the CpG islands in the IL-4 and -6 promoters. To test this hypothesis, we characterized the methylation pattern of the IL-4 and -6 promoters in activated lupus T cells. As showed in Figure 3C, the levels of the unmethylated CpG pairs in the IL-4 promoter increased significantly in activated lupus T cells, except at position –419, similar to that of the 5-azaC-treated T cells. Parallelingly, the levels of unmethylated CpG islands in the IL-6 promoter, except at position –468 of lupus T cells, were significantly higher than that of the control T cells (Figure 3A). The average levels of the unmethylated CpG in the IL-4 (0.24± 0.09 vs 0.09±0.04) or -6 (0.32±0.11 vs 0.12±0.05) promoter significantly increased in activated lupus T cells, as compared with that of the control T cells (P<0.05 for both comparisons; Figure 6A,6C). Importantly, the levels of unmethylated CpG in the IL-4 or -6 promoters were positively correlated to the mRNA levels of lupus patients (Figure 6B,6D, P=0.049 or P=0.005, respectively), as determined by logistic regression analysis. These data suggest that during the SLE process, spontaneous demethylation in the promoter of some cytokines may result in the elevated expression of cytokines and many pathogenic T cells, which can cause pathogenic T-cell hyperactivation.
Discussion
IL-4 and -6 are important cytokines promoting B-cell differentiation, and are associated with the pathogenesis of SLE. Importantly, high levels of IL-6 mRNA and protein were detected in freshly isolated lymphocytes from SLE patients, and high serum levels of IL-6 are correlated with disease activity and severity. To characterize the levels of these cytokines in Chinese SLE patients, we found that the levels of IL-4 mRNA transcripts in activated T cells were significantly higher than that of healthy controls. Our data were in disagreement with the findings of low levels of IL-4 in Caucasian SLE patients, which was similar to that in the healthy controls[20–22]. The differential levels of IL-4 expressed by activated T cells may be attributed to different genetic backgrounds among SLE patients. Furthermore, the levels of IL-6 mRNA transcripts were elevated significantly in activated T cells from SLE patients, as compared with that in controls, similar to that of other reports[23,24]. High levels of IL-4 and -6 expressions may contribute to the pathogenic process in Chinese SLE patients.
We have known that the methylation of DNA and the enzymes regulating methylation are crucial for the regulation of gene expression and contribute to understanding the pathogenesis of autoimmune diseases[1–3]. To understand the mechanism(s) underlying the overexpression of IL-4 and -6 in SLE T cells, we found that the treatment of activated healthy T cells with DNMT inhibitor 5-azaC significantly enhanced the mRNA transcription of IL-4 and -6. An analysis of methylation in the CpG islands of the IL-4 and -6 promoters revealed that the treatment of activated healthy T cells with 5-azaC induced a higher frequency of demethylation in the CpG islands, as compared with that of untreated control T cells. Therefore, demethylation of the CpG islands in the IL-4 and -6 promoters may mediate the overexpression of these cytokines in 5-azaC-treated T cells. Previous studies have shown that cloned Th2 cells treated with 5-azaC became autoreactive, responding to the major histocompatibility complex (MHC) molecule, but independent of specific antigen and the overexpression of LFA-1, and induced transferable systemic autoimmune diseases[25,26]. This new acquirement of autoimmunity stems from demethylation and the subsequent overexpression of diseases-related genes. Thus, the treatment of activated T cells with DNA methyltransferase inhibitors can remodel the chromatin structure, modify gene expression, and induce autoreactivity.
The higher levels of IL-4 and -6 mRNA transcriptions in activated T cells may also be as a result of the hypomethylation of these genes in the SLE patients. The characterization of the methylation status of the CpG islands in the IL-4 or -6 promoter showed that the levels of unmethylated CpG islands of the IL-4 or -6 promoter were significantly higher in SLE T cells than that in control T cells, particularly in the CpG islands near to the site where transcription starts. The higher levels of CpG demethylation in the IL-4 and -6 promoters of T cells from SLE patients were unlikely to be caused by the in vitro activation with PHA, as the in vitro activation of T cells from healthy controls did not increase the demethylation levels of the gene[27]. Importantly, the levels of unmethylation of the IL-4 or -6 promoter were significantly correlated to the SLEDAI scores in SLE patients. Therefore, the demethylation of these genes in T cells is likely to be a natural development during the pathogenic process. Given that decreased DNMT function, increased transcription of 2 methyl CpG-binding domain (MBD2 and MBD4) genes[28], hypomethylation of the genes of LFA-1, perforin, and CD70 are associated with abnormal T-cell function, autoreactive B lymphocytes[29], and human SLE development, our data provide new additional insights into understanding the mechanism underlying methylation-related regulation on gene expression and the pathogenesis of human SLE.
Conceivably, during the development of SLE, some unknown factors downregulate the function of DNMT and upregulate the expressions of MBD2 and MBD4, leading to the hypomethylation of many SLE-associated genes, such as LFA-1, perforin, CD70, and IL-4 and -6, leading to the overexpression of these genes. Subsequently, the overexpression of these genes promotes the activation and function of self-reactive T and B cells. These epigenetic alterations, together with the potential stimulation of B cells by hypomethylated mammalian DNA through the Toll-like receptor, mediate the development and progression of human SLE[30,31]. Notably, unlike permanent genetic alterations, epigenetic alterations usually are reversible. Several drugs, such as immunosuppressive medicines that are reminiscent of the signaling aberrations, reverse the pattern of epigenetic modifications and have already been used to treat SLE[32,33]. Other histone deacetylase inhibitors and DNA-demethylating drugs may also be a promising source of epigenetic therapies to treat SLE[32]. However, they may mediate gene-gene-specific hypermethylation. Therefore, a detailed analysis of DNA methylation will allow us to evaluate the value of DNA-demethylating drugs at the clinic.
In summary, we found that higher levels of IL-4 and -6 mRNA transcripts in T cells from Chinese SLE patients and the treatment of healthy activated T cells with 5-azaC induced higher levels of IL-4 and -6 expressions, which were associated with demethylation of the CpG islands of the IL-4 or -6 promoter in activated T cells. Importantly, an analysis of the methylation status of activated T cells revealed that the levels of unmethylated CpG in the IL-4 and -6 promoters were significantly elevated in SLE patients, as compared with that in the healthy controls. Finally, the hypomethylation levels of the IL-4 or -6 promoter were significantly correlated with the SLEDAI scores of patients. Therefore, these data suggest that many CpG islands in the IL-4 and -6 promoters in T cells undergo demethylation during the SLE process in vivo. The identification of hypomethylation in the CpG islands of the IL-4 and -6 promoters in activated SLE T cells provides additional insights into understanding the mechanism(s) underlying the pathogenesis of SLE. Potentially, our findings may provide a basis for the design of a new therapy aimed to revert epigenetic alternations for human SLE patients.
References
- Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin Immunol 2003;109:37-45.
- Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr 2002;132:S2401-5.
- Nazarenko SA. Impaired epigenetic gene activity regulation and human diseases. Vestn Ross Akad Med Nauk 2001;10:43-8.
- Turek-Plewa J, Jagodziñski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett 2005;10:631-47.
- Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet 2003;33:61-5.
- Cedar H. DNA methylation and gene activity. Cell 1988;53:3-4.
- Sekigawa I, Okada M, Ogasawara H, Kaneko H, Hishikawa T, Hashimoto H. DNA methylation in systemic lupus erythematosus. Lupus 2003;12:79-85.
- Januchowski R, Prokop J, Jagodziñski PP. Role of epigenetic DNA alterations in the pathogenesis of systemic lupus erythematosus. J Appl Genet 2004;45:237-48.
- Nijnik A, Ferry H, Lewis G, Rapsomaniki E, Leung JC, Daser A, et al. Spontaneous B cell hyperactivity in autoimmune-prone MRL mice. Int Immunol 2006;18:1127-37.
- Zhu J, Liu X, Xie C, Yan M, Yu Y, Sobel ES, et al. T cell hyperactivity in lupus as a consequence of hyperstimulatory antigen-presenting cells. J Clin Invest 2005;115:1869-78.
- Januchowski R, Wudarski M, Chwaliñska-Sadowska H, Jagodzinski PP. Prevalence of ZAP-70, LAT, SLP-76, and DNA methyltransferase 1 expression in CD4(+) T cells of patients with systemic lupus erythematosus. Clin Rheumatol 2008;27:21-7.
- Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum 1990;33:1665-73.
- Richardson B. DNA methylation and autoimmune disease. Clin Immunol 2003;109:72-9.
- Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum 2004;50:1850-60.
- Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, et al. DNA methylation and chromatin structure regulate T cell perforin gene expression. J Immunol 2003;170:5124-32.
- Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood 2002;99:4503-8.
- Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest 1993;92:38-53.
- Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol 1995;154:3025-35.
- Andrews DF, Nemunaitis J, Tompkins C, Singer JW. Effect of 5-azacytidine on gene expression in marrow stromal cells. Mol Cell Biol 1989;9:2748-51.
- Gómez D, Correa PA, Gómez LM, Cadena J, Molina JF, Anaya JM. Th1/Th2 cytokines in patients with systemic lupus erythematosus: is tumor necrosis factor alpha protective? Semin Arthritis Rheum 2004;33:404-13.
- Linker-Israeli M, Honda M, Nand R, Mandyam R, Mengesha E, Wallace DJ, et al. Exogenous IL-10 and IL-4 down-regulate IL-6 production by SLE-derived PBMC. Clin Immunol 1999;91:6-16.
- Csiszár A, Nagy G, Gergely P, Pozsonyi T, Pócsik E. Increased interferon-gamma (IFN-gamma), IL-10 and decreased IL-4 mRNA expression in peripheral blood mononuclear cells (PBMC) from patients with systemic lupus erythematosus (SLE). Clin Exp Immunol 2000;122:464-70.
- Linker-Israeli M, Wallace DJ, Prehn J, Michael D, Honda M, Taylor KD, et al. Association of IL-6 gene alleles with systemic lupus erythematosus (SLE) and with elevated IL-6 expression. Genes Immun 1999;1:45-52.
- McMurray RW, Hoffman RW, Nelson W, Walker SE. Cytokine mRNA expression in the B/W mouse model of systemic lupus erythematosus—analyses of strain, gender, and age effects. Clin Immunol Immunopathol 1997;84:260-8.
- Yung R, Powers D, Johnson K, Amento E, Carr D, Laing T, et al. Mechanisms of drug-induced lupus. II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupuslike disease in syngeneic mice. J Clin Invest 1996;97:2866-71.
- Richardson B, Ray D, Yung R. Murine models of lupus induced by hypomethylated T cells. Methods Mol Med 2004;102:285-94.
- Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum 2002;46:1282-91.
- Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Vilardell-Tarrés M. Transcript overexpression of the MBD2 and MBD4 genes in CD4+ T cells from systemic lupus erythematosus patients. J Leukoc Biol 2007;81:1609-16.
- Sekigawa I, Kawasaki M, Ogasawara H, Kaneda K, Kaneko H, Takasaki Y, et al. DNA methylation: its contribution to systemic lupus erythematosus. Clin Exp Med 2006;6:99-106.
- McCubbin MD, Hou G, Abrams GD, Dick R, Zhang Z, Brewer GJ. Tetrathiomolybdate is effective in a mouse model of arthritis. J Rheumatol 2006;33:2501-6.
- Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med 2005;202:1131-9.
- Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin–IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 2002;416:603-7.
- Ballestar E, Esteller M, Richardson BC. The epigenetic face of systemic lupus erythematosus. J Immunol 2006;176:7143-7.
