Nosocomial colonization due to imipenem-resistant Pseudomonas aeruginosa epidemiologically linked to breast milk feeding in a neonatal intensive care unit
Introduction
Pseudomonas aeruginosa is one of the main organisms responsible for drug-resistant health care-associated infections[1–3]. Its ability to survive and replicate in hospital in a wide range of ecological niches, sometimes hostile to other microorganisms, may create tenacious environmental reservoirs, such as faucets, tap water, sinks, respiratory-therapy equipment and even detergents and disinfectants[4,5]. Well known as a multiresistant organism, it is posing new therapeutic and public health challenges because of the emergence and widespread dissemination of strains producing metallo-β-lactamases (MBL). Indeed, these enzymes can confer resistance to all β-lactams except monobactams and are not susceptible to the clinically available β-lactamase inhibitors[3,6]. Several types of these enzymes have been detected; namely, IMP, VIM, SPM, GIM and SIM, mostly in P aeruginosa and other Gram-negative nonfermenters[7]. The IMP- and VIM-type enzymes are currently the most widespread and in Europe, where they were first detected, the VIM-type enzymes are overall the most prevalent[8,9]. Colonization and infection by P aeruginosa in premature neonates rise commonly from an environmental source, though health-care workers have proved to be involved as the reservoir or vehicle of transmission[10,11]. Preterm neonates are potentially at higher risk for infection with P aeruginosa because they may be immunocompromised, often undergo invasive procedures and require prolonged hospitalization[1,2,10,12]. Outbreaks of illness traced to contaminated feeding in neonatal intensive care units (NICU) are also increasingly reported in association with formula products and, though less frequently, expressed human milk[13,14].
We describe a one-year investigation of colonization by imipenem-resistant, MBL-producing P aeruginosa in the NICU of the Dipartimento Materno-Infantile at the University Hospital of Palermo, Italy and the study of cross-transmission and risk factors for colonization with this pathogen in the NICU.
Materials and methods
Setting This study was part of a larger prospective study that examined surveillance cultures in the NICU of the University Hospital of Palermo, Italy, from 2003 January 7 to 2004 January 6[12]. The unit is a section of the Dipartimento Materno-Infantile, which is also a referral centre for congenital malformation in Sicily. The department includes units of infertility and assisted reproduction and maternal-fetal medicine.
Approximately 200 neonates are admitted annually in the NICU under study. The NICU is divided into 2 rooms connected by sliding doors. Hand washing facilities are available in each room. Use of a liquid detergent with 4% chlorexidine gluconate is strictly enforced for all staff and visitors. Gloves are used routinely in aseptic procedures.
During the study period, the average patient-to-nurse ratio was 3:1 and 2:1 in the intermediate care and intensive care section, respectively. Prematurity was the most common reason for admission to the NICU (approximately 70%). Other diagnoses for admission included respiratory distress, congenital malformation, surgical conditions and infections.
Whenever possible, neonates were exclusively or predominantly fed human milk. Alternatively, sterile ready-to-feed formula was preferred, but some specialized formulas were also used and prepared from powdered formula products. In the case of expressed milk feeding, mothers were given a reusable personal collection kit to use with the breast pump. Milk was then chilled and transported to the hospital, where it was labeled and refrigerated until use within a maximum of 48 h. Warming in water bath and handling of breast milk before feeding was carried out in a designated area in the formula preparation room. As the first-line therapy for suspected early-onset sepsis a combination of ampicillin-sulbactam plus gentamicin was adopted.
Patients All neonates admitted during the period under study, who remained hospitalized for at least 48 h in the NICU and from whom at least one rectal swab culture could be obtained, were enrolled.
The following parameters were recorded for all neonates: demographic characteristics, gestational age, birthweight, inborn or outborn condition, delivery type, APGAR score, length of stay, co-morbidity conditions. During the NICU stay, data about use and duration of the following devices and procedures, that could be considered as possible risk factors were also collected: antimicrobial therapy, central venous and peripheral catheterization, nasogastric and endotracheal tube insertion, and type of feeding (ie, parenteral, breast milk or formula).
A rectal swab culture yielding Gram-negative bacilli in the absence of clinical symptoms and signs was considered evidence of colonization. Nosocomial infection was identified according to the recommendations of the Centers for Diseases Control, Atlanta, GA, USA[15]. Congenital anomalies were defined as conditions listed under the International Classification of Disease (ICD-9) codes 740 to 759. The study protocol was reviewed and approved by the Ethical Committee of the University of Palermo, and a written parental consent was obtained for each neonate.
Microbiological surveillance Rectal swabs were collected twice a week (every Tuesday and Friday) from all neonates throughout their NICU stay. Isolation and identification of P aeruginosa and other Gram-negative organisms were carried out as previously described[12]. After biochemical identification by API20E or API20NE (BioMerieux, Marcy-l’Etoile, France), susceptibility to a panel of 10 antimicrobial substances was assessed by disk diffusion on Mueller-Hinton agar plates, according to the Clinical Laboratory Standards Institute (CLSI) guidelines[16]: ampicillin (30 µg), amoxicillin-clavulanic acid (30 µg), carbenicillin (10 µg), cefotaxime (30 µg), ceftazidime (30 μg), ceftriaxone (30 µg), cefepime (30 µg), imipenem (30 µg), aztreonam (30 µg), gentamicin (10 µg). For the purpose of the study, a Gram-negative organism resistant to at least 3 different groups of antimicrobial agents (penicillins, cephalosporins, aminoglycosides, carbapenems) was defined as multidrug-resistant Gram-negative (MDRGN).
Metallo-β-lactamase production by imipenem-resistant strains of P aeruginosa was detected by the EDTA-imipenem disk synergy test[17]. PCR amplification was carried out by using oligonucleotide primers targeting conserved regions of blaVIM and blaIMP[18]. Sequencing of VIM genes was carried out as previously described[18].
Pulsed field gel electrophoresis (PFGE) was carried out on all available isolates of P aeruginosa. Chromosomal DNA was digested using XbaI endonuclease and resolved by PFGE in a CHEF-Mapper apparatus (Bio-Rad, Hercules, CA, USA). Strains were interpreted as indistinguishable, closely related, possibly related or unrelated strains according to the criteria of Tenover et al[19].
Statistical analysis The association between potential risk factors and colonization by imipenem-resistant, imipenem-susceptible P aeruginosa strains and other MDRGN organisms was analyzed for variables present at admission and during the NICU stay. Time dependent variables, (ie length of stay and of antimicrobial use), were measured as between admission and the first isolation of a MDRGN organism.
One-way ANOVA and Kruskal-Wallis statistic test were used for parametric and non parametric analysis, respectively, to evaluate differences between the variables considered. The relative risk (RR) and the 95% confidence intervals (CI) for the risk factors were also calculated. The association was tested by Pearson’s χ2 test and Fisher exact test for frequency analysis. A Cox proportional hazards regression model was used to identify risk factors associated with colonization by imipenem-resistant P aeruginosa. Confounder selection included variables significant at P≤0.10 in the preliminary analysis. For the purpose of this analysis, birthweight was stratified in 3 categories: ≤1500 g, 1501–2500 g, and >2500 g. Length of stay was also stratified in three categories: ≤1, 2, and >2 weeks, respectively. All P-values were two-sided and P-values less than 0.05 were considered statistically significant. Data were analyzed by the Epi Info software (version 6.0, Centers for Disease Control and Prevention, Atlanta, GA, USA) and SPSS Software (version 14.0, SPSS Inc, Chicago, IL, USA)
Results
During the study period, a total of 221 neonates were admitted to the NICU for at least 48 h. Among these, 11 could not be submitted to rectal swab cultures and were excluded from further analysis.
A total of 1021 rectal swab cultures were sequentially obtained from 210 neonates. The characteristics of the study population are summarized in Table 1. The average length of stay was 22.3 d (median 13 d, range 3–140). Ninety-two (43.8%) neonates received antibiotics during the NICU stay. Criteria for infection were met by 25 (11.9%) neonates, of whom 2 died.
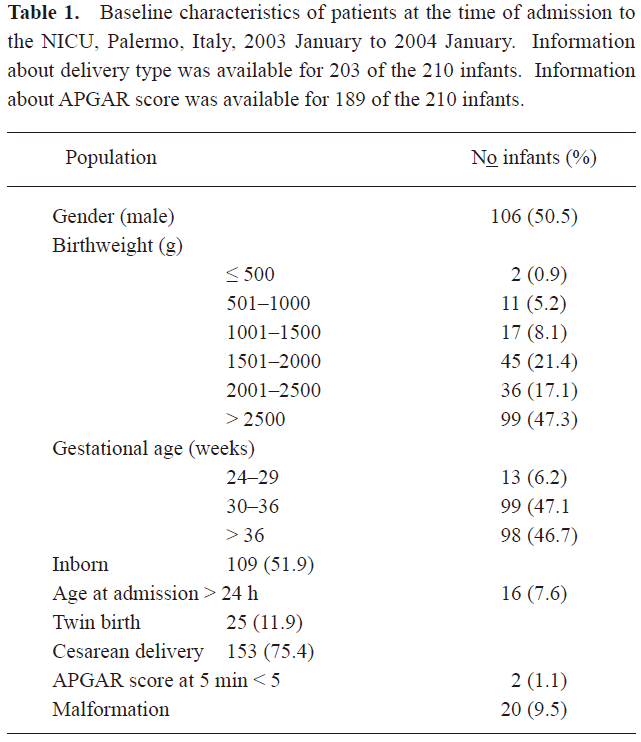
Full table
One-hundred and sixteen (55.2%) of 210 subjects were colonized with MDRGN at some point over the study period. The most frequent MDRGN species identified were Enterobacter cloacae (35.3%), Pseudomonas aeruginosa (31.0%), Klebsiella oxytoca (21.6%), Escherichia coli (19%), K pneumoniae (18.1%) and Serratia marcescens (7.8%). In particular, 22 neonates were colonized with an imipenem-resistant, MBL-producing P aeruginosa strain and 14 by an imipenem-susceptible P aeruginosa strain. No infection was detected in neonates colonized by these organisms.
Screening test of the MBL-producing strain of P aeruginosa gave positive PCR results with the primers for blaVIM genes. By sequence analysis, the strain was found to carry a novel variant of the MBL blaVIM family, designated blaVIM-11 (GenBank accession number AY635904).
The monthly incidence of acquisition of MDRGN, imipenem-resistant and imipenem-susceptible P aeruginosa, respectively, over the 12-month study period is illustrated in Figure 1. Strain relatedness of the P aeruginosa isolates was determined by PFGE analysis (Figure 2). A single pulsotype, named A, was shared by all imipenem-resistant P aeruginosa isolates, suggesting cross-transmission throughout the study period. On the contrary, the 14 imipenem-susceptible isolates were attributed to 11 different pulsotypes, named B to L. Only two were shared by 2 and 3 isolates from different neonates, respectively, whereas the remaining ones were unique.
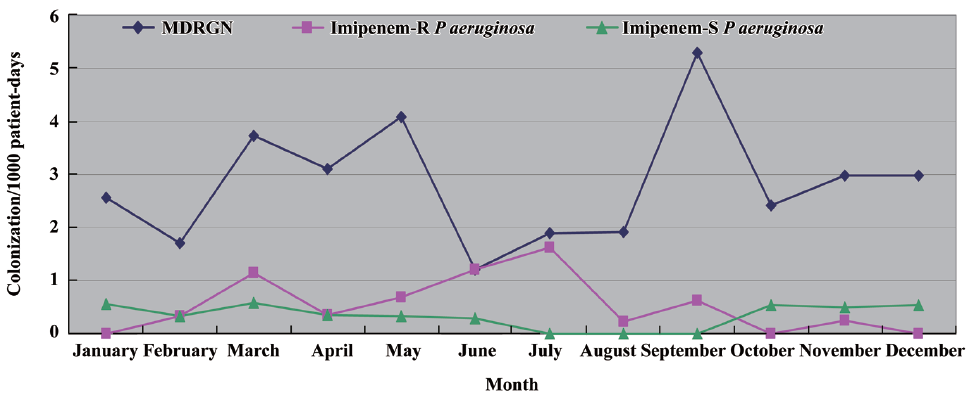
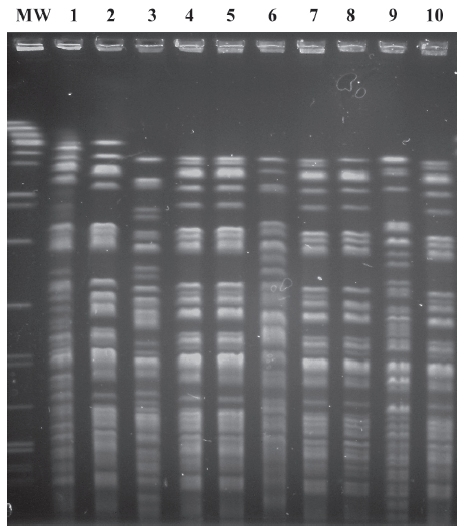
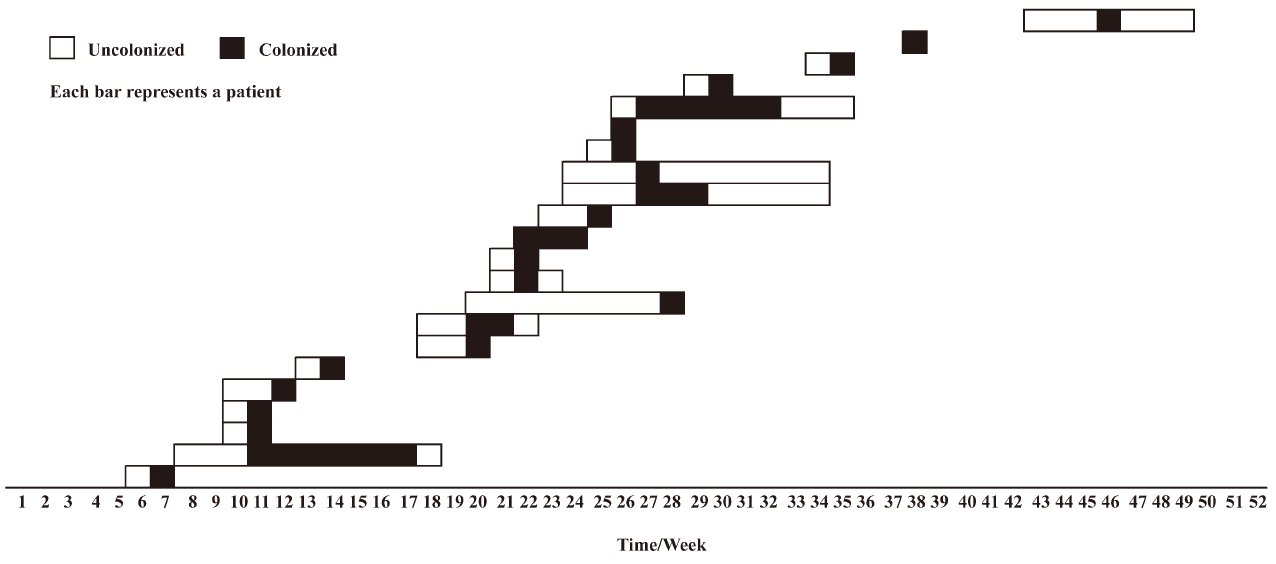
Figure 3 shows the pattern of transmission of P aeruginosa strains of pulsotype A among neonates during the 52 weeks of the period under investigation. A stepwise pattern was evident, but in some weeks the strain was not isolated, suggesting the possible contribution of an environmental reservoir to the persistence of the strain in the NICU.
We looked at the influence of selected clinical parameters at admission and during the NICU stay for their association with colonization with at least one strain of MDRGN, with imipenem-resistant, MBL-producing and imipenem-susceptible P aeruginosa strains. The association was initially tested by univariate analysis (Table 2). The only characteristic at admission significantly associated with colonization by P aeruginosa was a higher birthweight (Table 2). Furthermore, the NICU procedures most strongly associated with the acquisition of a colonization by MBL-producing P aeruginosa proved to be infant feeding, length of stay and administration of ampicillin-sulbactam (Table 2). Of interest, all of these characteristics, except for birthweight and length of stay, significantly differed between neonates colonized by imipenem-resistant and imipenem-susceptible isolates of P aeruginosa (Table 2).
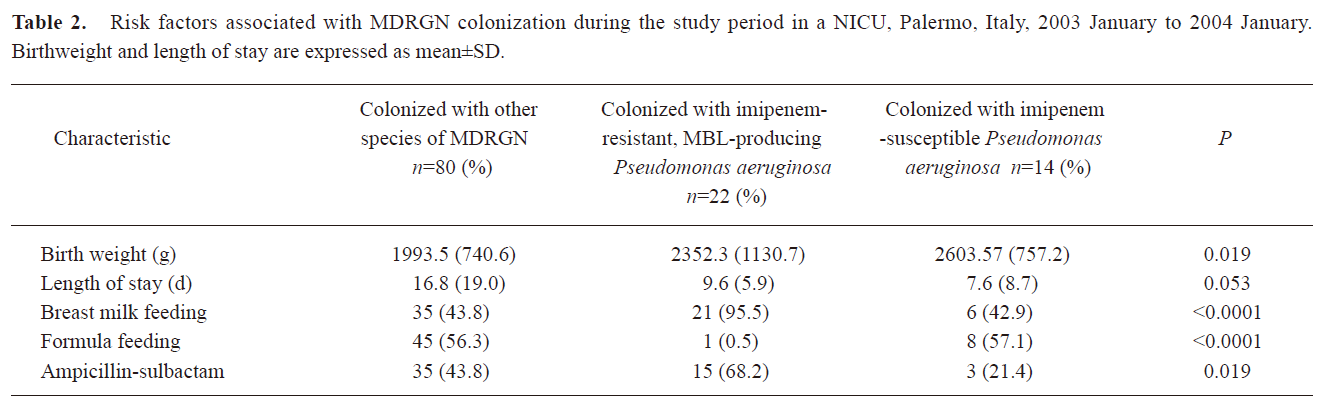
Full table
The risk of colonization in neonates positive for P aeruginosa of pulsotype A compared with those culturing positive for other multiresistant Gram-negatives, including imipenem-susceptible P aeruginosa, was positively correlated with breast milk feeding (RR=1.6, 95% CI, 1.3–1.9; P<0.001) and administration of ampicillin-sulbactam (RR=1.2, 95% CI, 1.1–1.5; P=0.01), and inversely correlated with exclusive feeding by formula (RR=0.7, 95% CI, 0.6–0.8; P<0.001). The administration of gentamicin (RR=1.1, 95% CI, 0.9–1.4; P=0.06) was not associated.
The significant variables at P 0.10 were further analyzed in the Cox regression model. A summary of results is provided in Table 3. In this model, birthweight of more than 2500 g (HR, 3.66; 95% CI, 1.21–11.13) and breast milk feeding (HR, 29.16; 95% CI, 3.91–217.69) were independently associated with an increased risk of colonization by MBL-producing P aeruginosa, whereas length of stay of more than 2 weeks (HR, 0.10; 95% CI, 0.00–0.11) and exclusive feeding by formula (HR, 0.18; 95% CI, 0.05–0.61) were inversely associated.
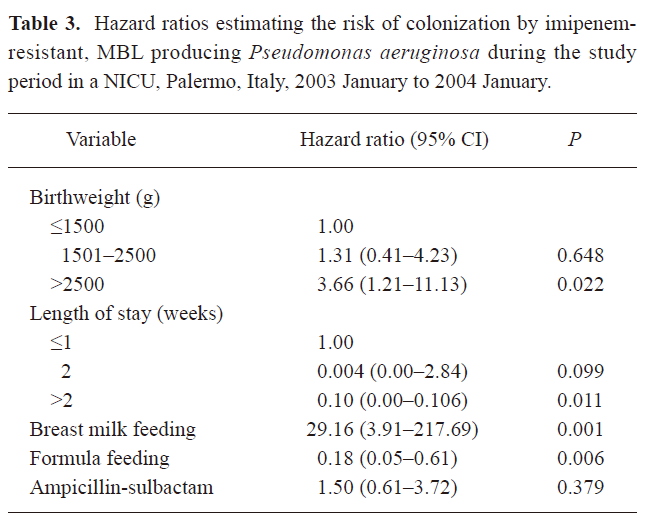
Full table
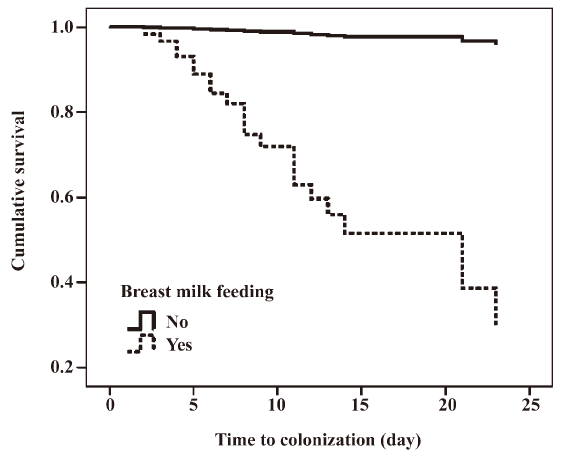
A survival curve comparing breast milk feeding status and number of days to first imipenem-resistant, MBL-producing P aeruginosa isolation is shown in Figure 4.
Discussion
The present study describes the transmission among 22 neonates admitted to the NICU throughout 12 months of a strain of imipenem-resistant P aeruginosa producing a novel blaVIM sequence. Colonization and infection by P aeruginosa in hospitalized neonates has been previously described[1,2,10]. However, the drug resistance pattern of the strain detected in our investigation is of special concern, because of the threat to the clinical utility of carbapenems, important therapeutic agents for the treatment of severe infections with MDRGN, particularly those carrying genes for extended-spectrum β-lactamases[7]. Although this organism was detected by surveillance cultures, we detected no infections in colonized neonates. Thus, its presence in the NICU would normally have been undetected.
As in previous studies, molecular typing by PFGE proved to be an invaluable tool to discriminate unique from shared strains and trace transmission routes. In particular, P aeruginosa isolates identified in the NICU were clearly differentiated in two groups: indistinguishable imipemem-resistant and MBL-producing isolates, an epidemiological synonym of clonality and cross-transmission, and highly heterogeneous imipenem-susceptible isolates, with only 2 micro-clusters of colonization cases.
In this study we found an association between colonization by a well-defined imipenem-resistant, MBL producing P aeruginosa strain and breast milk feeding. Contamination of human breast milk by P aeruginosa in NICU has been described in published reports[13,14]. Of particular interest, an outbreak in France in 2001 was caused by contamination of a milk bank pasteuriser and bottle warmer[13]. On the other hand, appropriate preparation and handling of formula for neonates in health-care facilities has received great attention since some years, whereas safe handling of human milk has been focused on only more recently[20]. High risk neonates are often unable to directly breastfeed. In these cases, mechanical expression, use of collection kits, cold storage, transport, and warming need to be carried out with techniques adequate to prevent contamination of milk[21]. Accordingly, guidelines of the American Dietetic Association have recently been updated, also including new chapters on handling of expressed human milk[20].
The pattern of spread documents some periods of cross-transmission, when P aeruginosa pulsotype A was recovered from neonates hospitalized during overlapping intervals, alternating with apparent eclipsing phases of the strain in the NICU. In many outbreak and non-outbreak contexts, the principal reservoir of MDRGN in NICU has proved to be the intestinal tract of the neonates and contaminated hands of healthcare workers the major vehicle of the continued dissemination of cross-colonizing strains[10,11]. Analytical limits of the detection method and timing of surveillance cultures might account for the holes in the transmission chain during the 12 months of observation. However, the apparent interruptions in the transmission route of the P aeruginosa strain could have been likely overcome by some environmental reservoir responsible for survival and perpetuation of the strain in NICU. Recovery of Gram-negative from inanimate surfaces, patient care equipment, contaminated fluids, water sources and water reservoirs has been repeatedly reported, with special reference to P aeruginosa[1,5]. Examination of formula, milk handling room and environmental flora was beyond the purposes of the study and was not carried out during the period under study. Therefore, association between the endemic imipenem-resistant P aeruginosa strain and the breastfeeding handling chain was based on epidemiological, but not microbiological, evidence. The results of the study were presented to the NICU staff and resulted in more emphasis on infection control measures; mainly hand washing, strict separation of clean and dirty procedures, attention to the general hygiene of rooms and equipment and safer management of all feeding-related aspects. Moreover, possible environmental reservoirs were sanitized or eliminated. Follow-up surveillance periods have been repeatedly conducted, and have documented the interruption of the transmission chain.
This surveillance study identified a specific practice likely responsible for the sustained circulation of imipenem-resistant, MBL producing P aeruginosa in the NICU under investigation. The ability to specifically detect circulation of clinically relevant strains through surveillance, timely hygienic interventions and cooperation by NICU personnel are crucial to minimize or control health care-associated infections.
As colonization by a multidrug-resistant organism may lead to serious consequences in NICU, such a study may highlight the need for development and application of strategies to prevent an invaluable nutrition support from becoming an insidious risk factor for acquisition of health care-associated infections.
Acknowledgements
The authors thank the health-care workers of the NICU where the study was conducted for their collaboration.
References
- Grundmann H, Kropec A, Hartung D, Berner R, Daschner F. Pseudomonas aeruginosa in a neonatal intensive care unit: reservoirs and ecology of the nosocomial pathogen. J Infect Dis 1993;168:943-7.
- Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, et al. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob Agents Chemother 1996;40:349-53.
- Zafar AB, Sylvester LK, Beidas SO. Pseudomonas aeruginosa infections in a neonatal intensive care unit. Am J Infect Control 2002;30:425-9.
- Blanc DS, Nahimana I, Petignat C, Wenger A, Bille J, Francioli P. Faucets as a reservoir of endemic Pseudomonas aeruginosa colonization/infections in intensive care units. Intensive Care Med 2004;30:1964-8.
- Trautmann M, Michalsky T, Wiedeck H, Radosavljevic V, Ruhnke M. Tap water colonization with Pseudomonas aeruginosa in a surgical intensive care unit (ICU) and relation to Pseudomonas infections of ICU patients. Infect Control Hosp Epidemiol 2001;22:49-52.
- Lagatolla C, Edalucci E, Dolzani L, Riccio ML, De Luca F, Medessi E, et al. Molecular evolution of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity. J Clin Microbiol 2006;44:2348-53.
- Walsh TR. The emergence and implications of metallo-beta-lactamases in Gram-negative bacteria. Clin Microbiol Infect 2005;11 Suppl 6:2-9.
- Cornaglia G, Mazzariol A, Lauretti L, Rossolini GM, Fontana R. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-beta-lactamase. Clin Infect Dis 2000;31:1119-25.
- Corvec S, Poirel L, Decousser JW, Allouch PY, Drugeon H, Nordmann P. Emergence of carbapenem-hydrolysing metallo-beta-lactamase VIM-1 in Pseudomonas aeruginosa isolates in France. Clin Microbiol Infect 2006;12:941-2.
- Foca M, Jakob K, Whittier S, Della Latta P, Factor S, Rubenstein D, et al. Endemic Pseudomonas aeruginosa infection in a neonatal intensive care unit. New Engl J Med 2000;343:695-700.
- Widmer AF, Wenzel RP, Trilla A, Bale MJ, Jones RN, Doebbeling BN. Outbreak of Pseudomonas aeruginosa infections in a surgical intensive care unit: probable transmission via hands of a health care worker. Clin Infect Dis 1993;16:372-6.
- Mammina C, Di Carlo P, Cipolla D, Giuffrè M, Casuccio A, Di Gaetano V, et al. Surveillance of multidrug-resistant Gram-negative bacilli in a neonatal intensive care unit: prominent role of cross-transmission. Am J Infect Control 2007;35:222-30.
- Gras-Le Guen C, Lepelletier D, Debillon T, Gournay V, Espaze E, Roze JC. Contamination of a milk bank pasteuriser causing a Pseudomonas aeruginosa outbreak in a neonatal intensive care unit. Arch Dis Child Fetal Neonatal Ed 2003;88:434-5.
- Thom AR, Cole AP, Watrasiewicz K. Pseudomonas aeruginosa infection in a neonatal nursery, possibly transmitted by a breast-milk pump. Lancet 1970;7646:560-1.
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections Am J Infect Control 1988;16:128-40.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 15th International supplement. CLSI documente M100-S15. Wayne: CLSI; 2005.
- Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter strains. Clin Microbiol Infect 2001;7:88-91.
- Sader HS, Castanheira M, Mendes RE, Toleman M, Walsh TR, Jones RN. Dissemination and diversity of metallo-beta-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int J Antimicrob Agents 2005;25:57-61.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995;33:2233-9.
- American Dietetic Association. Pediatric Nutrition Practice Group: Infant feedings: guidelines for preparation of formula and breast milk in healthcare facilities. Chicago; 2004.
- Pejaver KR, Toonisi MA, Garg AK, al Hifzi I. Is expressed breast milk from home safe? A survey from a neonatal intensive-care unit. Infect Control Hosp Epidemiol 1996;17:346-7.
