Amantadine as a regulator of internal ribosome entry site1
Introduction
Translation initiation in eukaryotes proceeds in 2 ways: a cap- and 5′ end-dependent mechanism and a cap-independent mechanism that acts through an internal RNA element termed the internal ribosomal entry site (IRES). IRES elements were first discovered in the RNAs of the virus family Picornaviridae, which have long, highly structured 5′ untranslated regions (5′UTR) but lack a cap structure at the 5′ end[1,2]. In these viruses, the IRES can fold to be a functional secondary RNA structure and drives translation initiation, thereby the IRES can replace the function of some initiation protein factors, allowing cap-independent translation[3]. Viral proteases cleave protein translation initiation factors like eIF4G, reducing the efficiency of the host cell’s cap-dependent translation initiation and favoring the virus IRES-mediated translation.
Internal ribosomal entry site elements are not restricted to picornaviruses but have also been found in the genomes of retroviruses and DNA viruses, like HIV and herpes simplex virus[4]. More interestingly, IRES elements have also been found in several cellular mRNAs[5]. In eukaryotic cells, IRES-dependent translation of cellular mRNAs has been reported to occur when cap-dependent translation is impaired, for instance, under conditions of apoptosis, heat shock stress, viral infection, and in the G2/M phase of the cell cycle[5–7]. Further studies have also suggested that the IRES may increase translation efficiency at postsynaptic sites following synaptic activation[8].
In addition to being an unusual translational mechanism, IRES have been successfully introduced between 2 cistrons to construct bicistronic or polycistronic vectors[9,10], an application that has been important in biotechnology[11]. This bicistronic vector allows the simultaneous expression of 2 proteins: a selection marker (or a reporter gene) and another gene of interest. The use of IRES to mediate gene expression has several advantages: (a) it ensures the coexpression of the 2 genes in more than 90% of cells[10]; (b) there is a fixed expression ratio between the level of expression of the first cistron cloned upstream of the IRES and the expression level of the second cistron inserted downstream of the IRES[10,12]; (c) IRES can mediate translation from RNA lacking the 5′-terminal cap structure as produced by RNA polymerase I or III[13–16]; (d) IRES can be activated in situations where cap-dependent translation is impaired, for example, during apoptosis or certain stages of the cell cycle[4]. All of these characteristics have made IRES an important tool for genetic engineering.
At present, IRES elements from encephalomyocarditis virus (EMCV) are the most commonly used modules for the construction of bicistronic expression vectors[9]. However, EMCV IRES-mediated translation is less efficient than the cap-dependent translation of the upstream gene[17,18]. We have previously reported a potent IRES derived from the picornavirus Enterovirus 71 (EV71) and demonstrated the EV71 IRES translation efficiency to be approximately 10- to 15-fold higher than that of EMCV IRES in cell lines[19]. In this study, we identified amantadine, a compound that inhibited EV71 IRES as well as EMCV IRES-mediated translation but did not interfere with cap-dependent translation. Our results suggest that amantadine may distinguish between cap-dependent translation and cap-independent IRES-mediated translation and can be used to regulate gene expression at a translational level.
Materials and methods
Cell culture, plasmids and transfection COS-1 (African green monkey kidney fibroblast-like cell line) and N2A (a rat neuroblastoma cell line) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). We generated the plasmids pGS-EV71 and pGS-EMCV as described by Lee et al[19]. These contained IRES elements derived from EV71 and EMCV, respectively, flanked by the β-galactosidase as well as secreted alkaline phosphatase (SEAP) reporter genes. COS-1 cells were then transfected with the plasmids using the Lipofectin reagent (Invitrogen, Carlsbad, CA, USA) and plated onto 24-well plates at a density of 0.5×105−2×105 cells/well. The cells were repeatedly washed with serum-free media to remove all traces of sera prior to transfection. One microgram of plasmid was diluted in 200 µL of serum-free DMEM medium, and one microliter of the Lipofectin reagent was added. The DNA-Lipofectin mix was incubated for 15 min to allow DNA-Lipofectin complex formation and then transferred to the cells at a final volume of 0.5 mL in serum-free medium. The transfected cells were then incubated at 37 °C in 5% CO2. After 12 h, the transfection media were replaced by media containing FBS and antibiotics and cultured for a further 12 h before being used in the following compound screen.
Compound screen Candidate compounds were dissolved in phosphate-buffered saline (PBS) to yield a stock concentration of 10 mg/mL. The various diluted compound solutions were then added to the plasmid-transfected cells and incubated for 12 h. The cells were lysed and the reporter protein activities (ie, β-gal and SEAP) were measured to screen for compounds that inhibit IRES-dependent translation.
Measurements of SEAP and β-galactosidase activity Posttransfection, supernatants were harvested and analyzed for SEAP activity using BD Great EscApe SEAP detection kits (Clontech, Mountain View, CA). For the β-galactosidase activity assay, the transfected cells were lysed for 10 min in 300 µL of culture cell lysis reagent containing 100 mmol/L potassium phosphate (pH 7.8), 1 mmol/L EDTA, 10% Triton X-100, and 7 mmol/L β-mercaptoethanol. After centrifugation at 15 200×g for 30 min, the lysate supernatant was measured for β-galactosidase activity using a Luminescent β-galactosidase Detection Kit II (BD Biosciences, Franklin Lakes, NJ, USA). The chemiluminescent intensities reflecting relative SEAP activities and β-galactosidase activities were detected with a chemical luminescence counter (Mithras LB 940, Berthold Technologies). Both SEAP and β-galactosidase activity were expressed as relative light units (RLU). The detection limit SEAP enzyme was about 10–13 g. Thus, the sensitivity of our bicistronic assay is comparable with renilla and firefly-luciferase assay. Besides, SEAP is secreted into the culture medium; so we can measure the alkaline phosphatase activity without lysis of cells.
Results
Rationale of experimental design Although protein translation is evolutionarily conserved in both prokaryotic and eukaryotic cells, some antibiotics are able to distinguish translational differences between the 2 cell types and specifically target translation in one cell type. Indeed, the inhibition of prokaryotic protein synthesis by antibiotics such as hygromycin and tetracycline is a triumph of modern medicine[20]. Based on this, we hypothesized that it might be possible to identify compounds that may distinguish between cap-dependent translation and cap-independent IRES-mediated translation. A possible model for this mechanism is shown in Figure 1A. Accordingly, we sought to identify compounds that inhibit IRES-driven translation (ORF2) without affecting cap-dependent translation (ORF1).
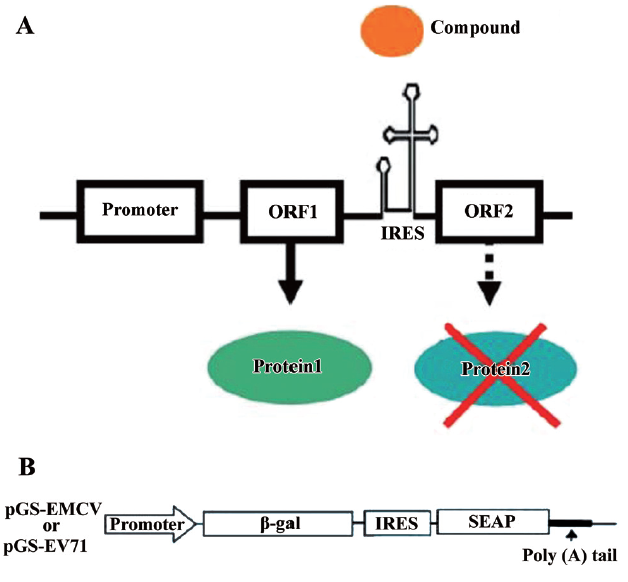
To facilitate compound screening, we developed bicistronic reporter constructs containing a β-galactosidase gene (β-gal) and a secreted human placental alkaline phosphatase (SEAP) reporter gene (Figure 1B). In these bicistronic expression vectors, a single construct containing the β-gal and SEAP reporter genes is under transcriptional control of the human cytomegalovirus major intermediate early promoter/enhancer sequence (CMV promoter). Following transcription, the β-gal gene is translated by a cap-dependent mechanism, while SEAP expression is controlled by either EV-71 IRES (pGS-EV71) or EMCV IRES (pGS-EMCV). This assay could potentially identify compounds that inhibit SEAP expression (cap-independent) without affecting β-gal activity (cap-dependent).
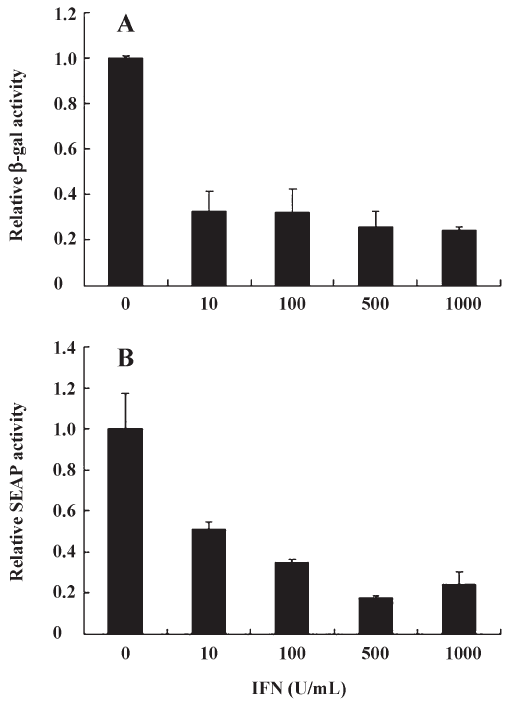
IFN-α inhibits both cap-dependent and cap-independent translation Interferon-alpha (IFN-α) combined with ribavirin is the most widely used treatment against hepatitis C virus (HCV), a flavivirus that initiates translation using IRES[21]. IFN-α acts by inhibiting HCV- and EMCV-IRES dependent translation[21]. Furthermore, The effect of IFN on translation control during virus infection has been extensively studied[22]. IFN is known to induce PKR, which increases phosphorylation levels of eIF2a, which in turn inactivates cap-dependent translation. Thus, we decided to validate our compound screen by testing the effects of IFN-α on EV71 IRES- as well as cap-dependent translation in pGS-EV71 transfected COS-1 cells. Although the levels of EV-71 IRES-mediated translation in pGS-EV71-transfected COS-1 cells decreased with the increasing concentration of IFN-α (Figure 2B), the levels of cap-dependent translation also decreased (Figure 2A). At 500 U/mL of IFN-α, the translational activity of both EV-71 IRES and cap were approximately 20% of the untreated cells (Figure 2). Since IFN-α efficiently suppressed the translational activity of EV-71 IRES but also interfered with the cap-dependent translation, it did not fit the criterion of inhibiting cap-independent translation without affecting cap-dependent translation.
Amantadine can inhibit IRES-mediated translation without affecting cap-dependent translation in low dose A compound identified in our screen that fit these criteria is amantadine. Amantadine, developed in the 1960s, has diverse uses that range from prevention of influenza A infection to the treatment of Parkinson’s disease[23]. This tricyclic symmetric amine can inhibit replication of myxoviruses, including influenza A viruses. It exhibits antiviral activity against influenza A through specific binding to the viral matrix (M2) protein[24]. Although sequence comparison shows no homology between M2 and any of the HCV proteins, amantadine has been reported to be an effective treatment in some patients with chronic HCV infection[25]. Recently, amantadine has been found to inhibit HCV IRES translation when used at above clinically-relevant concentrations[26]. Amantadine also suppresses HAV IRES translation[27]. However, in both reports, amantadine also suppressed cap-dependent translation activity, in a rabbit reticulocyte-based in vitro transcription/translation assay system or in human hepatoma-derived cell lines.
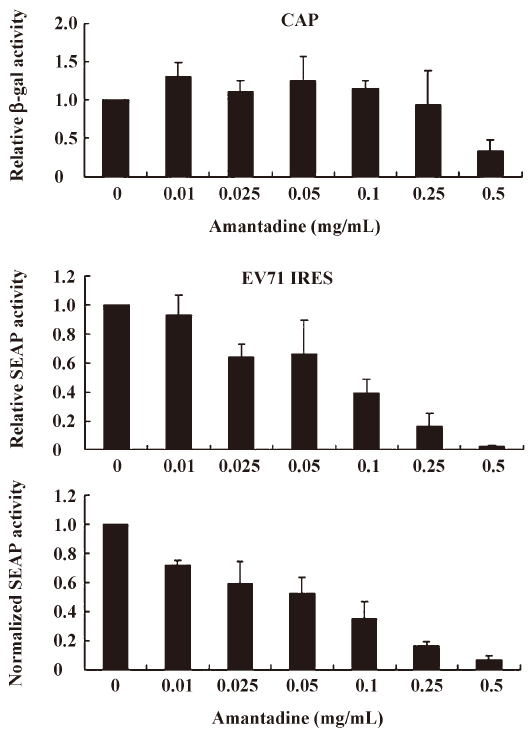
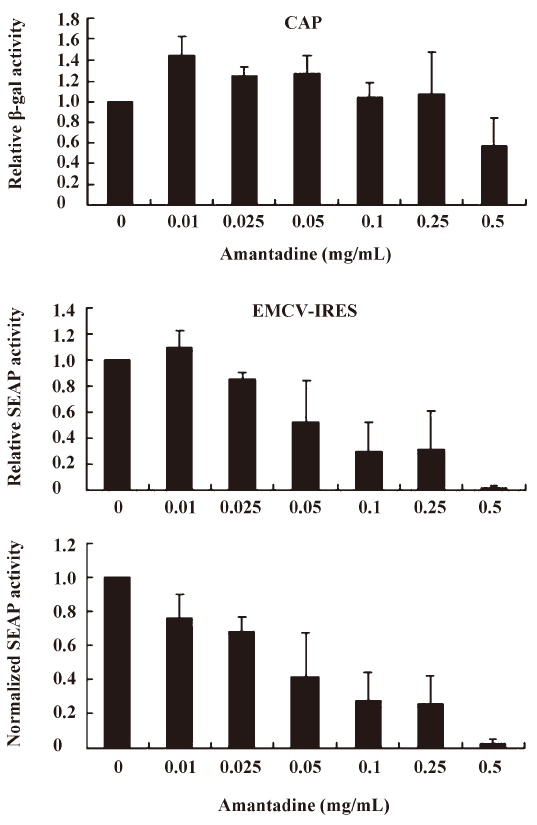
We found that incubating pGS-EV71-transfected COS-1 with amantadine (0.01 mg/mL to 0.25 mg/mL) for 12 h instead of 30 h, as reported by Kanda et al[27], had little or no effect on the β-gal activities (Figure 3, CAP). Interestingly, amantadine at these concentrations inhibits SEAP activities in a dose-dependent manner (Figure 3, EV71 IRES). However, both the β-gal activities and SEAP activities were inhibited by more than 50% if the transfected COS-1 cells were incubated with 0.5 mg/mL amantadine for 12 h (Figure 3). These results suggest that the EV71 IRES mediated translation is more sensitive than cap-dependent translation to amantadine. In particular, amantadine at a dose of 0.25 mg/mL inhibited about 80% of EV71 IRES-dependent translation activity with relatively little effect on CAP-dependent translation activity (Figure 3). Besides, we also demonstrated that the amantadine could also suppress the EV71 IRES mediated translation selectively in N2A cells (Figure 4). These results suggest that amantadine at a dose of less than 0.25 mg/mL fits our screening criteria for a compound that specifically inhibits EV71 IRES-mediated translation while leaving CAP-dependent translation unaffected.
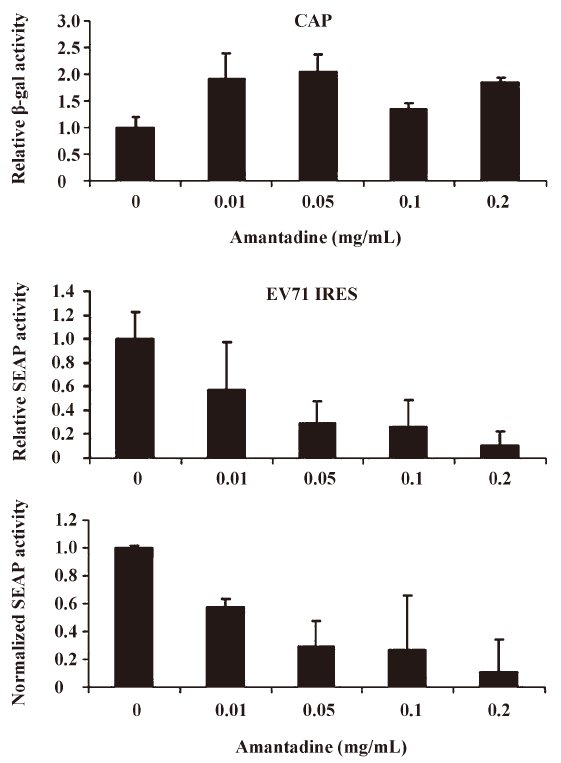
Amantadine inhibits both type I and type II class IRES-mediated translation Picornavirus IRES have been classified into 4 types according to their predicted secondary structure and in vitro activity[3]. Type I IRES elements are found in entroviruses and rhinoviruses, whereas type II elements are found in cardioviruses and aphthoviruses. Since the EV71 IRES belongs to the type I class, we were also interested to see if amantadine would also inhibit the type II class, like the EMCV IRES. We found that 0.01 mg/mL to 0.25 mg/mL amantadine specifically inhibited EMCV IRES-mediated SEAP translation but did not interfere with CAP-mediated β-gal translation (Figure 5). These results demonstrate that amantadine may inhibit types I and II IRES-mediated translation specifically under our assay conditions.
Discussion
In the present study, we identified amantadine as being able to inhibit EV71 IRES- as well as EMCV IRES-mediated translation without suppressing cap-dependent translation under our assay conditions. A potential application of this finding is to develop a system of regulating genes at a translational level by using amantadine to inhibit IRES-mediated translation.
The conventional gene inducible vector systems are usually comprised of 3 parts: (a) an inducible promoter such as a lac promoter[28] or a TRE-CMVmin promoter[29]; (b) a protein transcriptional activator or a repressor such as the tTa transactivator that activates the TRE-CMVmin promoter or the lacI repressor that suppresses the lac promoter; and (c) a regulator, usually a small molecule such as IPTG or tetracycline, that is capable of controlling the binding activities between the protein transcription activators or repressors and promoters. This transcription-based gene regulation system requires specific promoters and corresponding transcriptional factors as well as regulators.
Based on the findings of this study, a more simplified gene regulation system may be obtained by taking advantage of the unique properties of amantadine and bicistronic DNA constructs containing EV71 or EMCV IRES. We tentatively name this system the “IRES repressor.” With the IRES repressor, no specific promoters or protein factors are required. In addition, the bi-cistronic vector-based gene regulation system may simultaneously express a reporter gene or antibiotic-selection gene by cap-dependent translation for internal control of transfection or stable cell line selection, respectively. As shown in Figure 3, in an IRES switch system containing an EV71-IRES, a 20% gene inhibition was achieved by use of amantadine at a dose of about 0.01 mg/mL, whereas an 80% gene inhibition was achieved at a dose of about 0.25 mg/mL. Similarly, in the system containing an EMCV-IRES (Figure 4), a 20% or 80% gene inhibition was obtained at a dose of amantadine of about 0.05 mg/mL or 0.25 mg/mL, respectively. Interestingly, the expression of certain genes is controlled by the 5′UTR of mRNA that forms receptors called riboswitches for target metabolites. These riboswitches have been discovered in both Gram-positive and Gram-negative bacteria. Riboswitches, similarly to IRES, may form a specific RNA structure that acts as an aptamer or molecular switch in response to direct binding of diverse metabolites to regulate gene translation and control bacterial metabolites. The IRES switch, together with the riboswitch, provides evidence that genetic control may be manipulated post-transcription. Although the precise mechanism of IRES inhibition by amantadine remains unclear, amantadine may act similarly to riboswitches by selectively binding to specific IRES RNA conformation or factors that mediate the IRES-dependent translation, such as IRES trans-acting protein factors (ITAF), thereby blocking events in IRES-dependent translation. Further studies to clarify this will be of interest for the understanding of IRES mediated translation.
Acknowledgements
The authors thank the support from The Center of Excellence Program on Membrane Technology, the Ministry of Education of Taiwan.
References
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988;334:320-5.
- Jang SK, Krausslich HG, Nicklin MJH, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 1988;62:2636-43.
- Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol 2008;16:230-7.
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 2001;15:1593-612.
- Sachs AB. Cell cycle-dependent translation initiation: IRES elements prevail. Cell 2000;101:243-5.
- Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol 1999;1:190-2.
- Martinez-Salas E, Ramos R, Lafuente E, Lopez de Quinto S. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J Gen Virol 2001;82:973-84.
- Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci USA 2001;98:2770-5.
- Martinez-Salas E. Internal ribosome entry site biology and its use in expression vectors. Curr Opin Biotechnol 1999;10:458-64.
- de Felipe P. Polycistronic viral vectors. Curr Gene Ther 2002;2:355-78.
- Levenson VV, Transue EDE, Roninson IB. Internal ribosomal entry site-containing retroviral vectors with green fluorescent protein and drug resistance markers. Hum Gene Ther 1998;9:1233-6.
- Liu X, Constantinescu SN, Sun Y, Bogan JS, Hirsch D, Weinberg RA, et al. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal Biochem 2000;280:20-8.
- Elroy-Stein O, Fuerst TR, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5' sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci USA 1989;86:6126-30.
- Zhou Y, Giordano TJ, Durbin RK, McAllister WT. Synthesis of functional mRNA in mammalian cells by bacteriophage T3 RNA polymerase. Mol Cell Biol 1990;10:4529-37.
- Palmer TD, Miller AD, Reeder RH, McStay B. Efficient expression of a protein coding gene under the control of an RNA polymerase I promoter. Nucleic Acids Res 1993;21:3451-7.
- Royall E, Woolaway KE, Schacherl J, Kubick S, Belsham GJ, Roberts LO. The Rhopalosiphum padi virus 5' internal ribosome entry site is functional in Spodoptera frugiperda 21 cells and in their cell-free lysates: implications for the baculovirus expression system. J Gen Virol 2004;85:1565-9.
- Lee AH, Han JM, Sung YC. Generation of the replication competent human immunodeficiency virus type I which expresses a jellyfish green fluorescent protein. Biochem Biophys Res Commun 1997;233:288-92.
- Hennecke M, Kwissa M, Metzger KA, Oumard A, Kroger R, Schirmbeck J, et al. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res 2001;29:3327-34.
- Lee JC, Wu TY, Huang CF, Shih SR, Hsu JTA. High-efficiency protein expression mediated by enterovirus 71 internal ribosome entry site. Biotechnol Bioengineering 2005;90:656-72.
- Ochi K. From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem 2007;71:1373-86.
- Koev G, Duncan RF, Lai MMC. Hepatitis C virus IRES-dependent translation is insensitive to an eIF2α-independent mechanism of inhibition by interferon in hepatocyte cell lines. Virology 2002;297:195-202.
- Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci 2007;32:57-62.
- Aoki FY, Sitar DS. Clinical pharmacokinetics of amantadine hydrochloride. Clin Pharmacokinet 1988;14:35-51.
- Sugre RJ, Hay AJ. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology 1991;180:617-24.
- Smith JP. Treatment of chronic hepatitis C with amantadine. Dig Dis Sci 1997;42:1681-7.
- Jubin R, Murray MG, Howe AYM, Butkiewicz N, Hong Z, Lau JYN. Amantadine and rimantadine have no direct inhibitory effects against hepatitis C viral protease, helicase, ATPase, polymerase, and internal ribosomal entry site-mediated translation. J Infect Dis 2000;181:331-4.
- Kanda T, Yokosuka O, Imazeki F, Fujiwara K, Nagao K, Saisho H. Amantadine inhibits hepatitis A virus internal ribosomal entry site-mediated translation in human hepatoma cells. Biochem Biophys Res Commun 2005;331:621-9.
- Yarranton GT. Inducible vectors for expression in mammalian cells. Curr Opin Biotechnol 1992;3:506-11.
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 1992;89:5547-51.
