α-Helical domain is essential for antimicrobial activity of high mobility group nucleosomal binding domain 2 (HMGN2)1
Introduction
Cationic antimicrobial peptides play a crucial role in the host defense against infections[1]. It has been shown that the antimicrobial activity of human mononuclear leukocytes originates from natural killer (NK) cells[2], T cells[3] and monocytes[4]. Several antimicrobial peptides or proteins have been described in NK and T cells. Granulysin is a recently characterized antimicrobial polypeptide from T and NK cells[5]. The porcine counterpart of granulysin (NK-lysin) was characterized as an antimicrobial and cytotoxic polypeptide expressed by NK and T cells[6]. The antimicrobial peptides LL-37 and human neutrophil defensins (HNP1-HNP3) were found in the supernatant of enriched T and NK cells[7]. We isolated and characterized an antimicrobial polypeptide from human circulatory mononuclear leukocytes. Its N-terminal sequence was identical to high mobility group nucleosomal binding domain 2 (HMGN2)[8]. HMGN2 was identified as a non-histone chromosomal protein in invertebrates and vertebrates and may play a role in gene transcription and organogenesis[9–12]. However, the biological role of this protein has not been fully defined. In this study, we prepared recombinant holo-molecule, recombinant or synthetic helical domain, N-terminal and C-terminal fragments to further determine the antimicrobial spectrum and functional structure of HMGN2.
Materials and methods
Synthetic peptide Synthetic N- and C-terminal fragments, and the α-helical domain of HMGN2, were prepared by Shanghai Genebase Gen-Tech (Shanghai, China). Their amino acid sequences are as follows:
Fragment 1: MPKRKAEGDA KGDKAKV (position 1-17 of the HMG 2 amino acid sequence)
Fragment 2: KDEPQRRSAR LSAKPAPPKP EPKPKKAPAK K (position 18–48 of the HMGN2 amino acid sequence)
Fragment3: GEKVPKGKKG KADAGKEGNN PAENGDA-KTD QAQKAEGAGD AK (position 49–90 of the HMGN2 amino acid sequence)
High-performance liquid chromatography (HPLC) and mass spectroscopy analysis of these peptides revealed a purity of >95%. The peptides were dissolved in 10 mmol/L potassium phosphate buffer (PPB, pH 7.0) to a final concentration of 10 g/L.
Antimicrobial activity assay Rabbit neutrophil defensin (NP-1) and HNP1–3 were used as the control antimicrobial peptides, and were prepared as described elsewhere[13,14].
Evaluation of minimal effective concentration The minimal effective concentrations (MEC) were tested using a radial diffusion assay. Briefly, soy broth (E coli ML-35p, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 25923) or sabouraud dextrose broth (Candida albicans ATCC 10231) underlay gel mixture containing 1×106 colony-forming units (CFU)/mL of organisms was decanted into a dish. Sample wells of 3-mm diameter were punched and 5 μL of serial dilutions of the peptides (200, 100, 50, 25, 12.5, 6.25, and 3.125 mg/L) were added to the wells. After 3 h of incubation, overlay soy broth (for growing E coli ML-35p, P aeruginosa ATCC 27853, S aureus ATCC 25923) or sabouraud dextrose broth (for growing C albicans ATCC 10231) gels were poured and incubated continuously at 37 °C overnight, the resulting clear zones were measured and expressed in units (1 mm=10 U) after subtracting the well dia-meter. A linear regression analysis of peptide concentration (x-axis) against the zone diameter (y-axis) was carried pit to determine the x-intercept, whose value represented the MEC.
Evaluation of minimal inhibitory concentration and minimal bactericidal concentration The minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) of the peptides were examined using bacteria at 1×106 CFU/mL in the soy broth and serial dilutions of the peptides (500, 250, 200, 150, 100, 50, 25, 12.5, and 6.25 mg/L). Inhibition of growth was determined by measuring optical density (OD) at 492 nm on a UV/VIS spectrometer after incubation at 37 °C for 12–16 h. Antimicrobial activity was expressed as the MIC, the concentration at which 100% inhibition of growth was observed, and the MBC, the concentration at which no CFU were observed after incubation for 12–16 h on soy broth (for growing E coli ML-35p, P aeruginosa ATCC 27853, and S aureus ATCC 25923) or sabouraud dextrose broth (for growing C albicans ATCC 10231) solid media.
E coli-based production of recombinant human holo-HMGN2 and its α-helical domain Total RNA was isolated with Trizol Reagent (Gibco BRL, Washington, USA) from the stimulated mononuclear leukocytes. The full-length HMGN2 cDNA was amplified by reverse transcription–polymerase chain reaction (RT-PCR) and ligated to pMD-18T vector (TakaRa,Tokyo, Japan) for DNA sequencing. Generation of DNA of holo-HMGN2 and the HMGN2 α-helical domain was carried out by PCR amplification. Primers containing BamH I and EcoR I restriction sites were designed as follows: P1 (5'-AC
The transformed E coli JM109 carrying pGEX-1λT-HMGN2 and pGEX-1λT-HMGN2α were cultured in Luria-Bertani (LB) medium for 12 h in the presence of isopropylthio-β-D-galactoside (IPTG) to induce protein expression. The induced cultures were washed with phosphate-buffered saline and cell lysates were obtained by freezing/thawing in the presence of lysozyme. After centrifugation (10000 r/min, 4 ºC, 5 min), the fusion proteins were purified from the supernatants using Bulk Glutathione Sepharose 4B columns (Amersham Biosciences). The purified fusion proteins were cleaved by thrombin digestion. Holo-HMGN2 and its α-helical domain were obtained by acid-urea polyacrylamide gel electrtophoresis (AU-PAGE) elution and HPLC purifica-tion.
Results
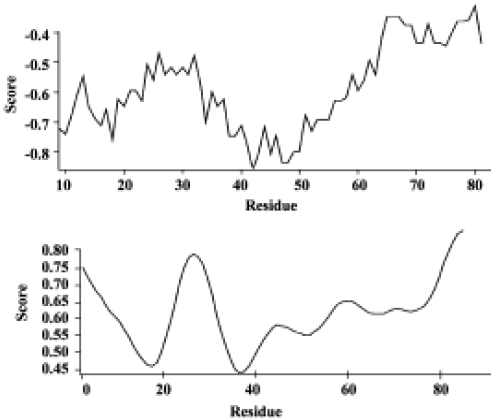
Analysis of the HMGN2 two-dimensional structure OMIGA protein structure software analysis revealed a transmembrane α-helical structure, the putative antimicrobial domain, located from position 18 to 48 of the HMGN2 protein sequence (Figure 1).
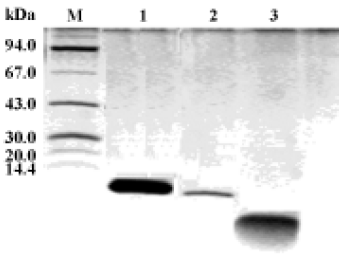
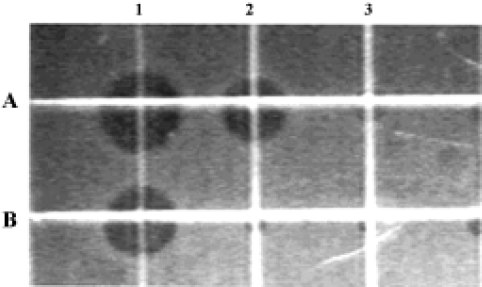
Antimicrobial activity of the synthetic peptide fragments Sodium dodecyl sulphate-polyacrylamide gel electrophoresis of the synthetic fragments of HMGN2 is shown in Figure 2. The agar radial diffusion assay indicated that the α-helical domain of HMGN2 had strong antimicrobial activity against an antibiotic-resistant strain of E coli. In contrast, no antimicrobial activity was observed for its N-terminal or C-terminal fragments using this assay system (Figure 3).
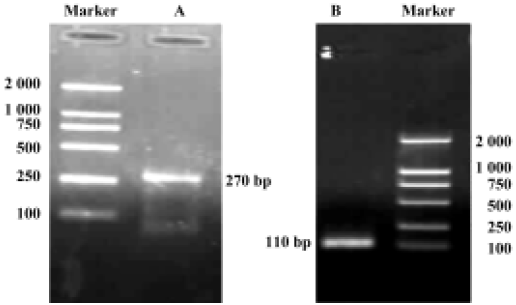
Construction of holo-HMGN2 and HMGN2 α-helical domain prokaryotic expression vectors The cDNA encoding mature holo-HMGN2 and its α-helical domain were obtained by PCR (Figure 4), and their corresponding prokaryotic expression vectors were constructed. Sequence analysis indicated that the insert sequences and their orientation were correct in the recombinant vectors.
E coli-based production of human holo-HMGN2 and its α-helical domain pGEX-1λT-HMGN2 or pGEX-1λT-HMGN2α-transformed E coli produced bulk amounts of the HMGN2 and HMGN2α fusion proteins. The fusion proteins were purified by glutathione S-transferase (GST) affinity chromatography. The purified recombinant holo-HMGN2 and its α-helical domain were obtained using AU-PAGE elution from thrombin-digested fusion proteins and reverse-phase HPLC (Figure 5).
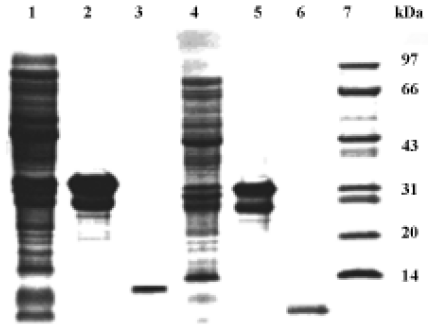
Antimicrobial properties of human holo-HMGN2 and its α-helical domain As shown in Table 1, the MIC, MEC, and MBC assays indicated that the recombinant human HMGN2 and its transmembrane α-helical domain (synthetic and recombinant) had potent antimicrobial activity against E coli, P aeruginosa and, to some extent, against C albicans. However, human HMGN2 was inactive against S aureus in this assay system (data not shown). All experiments were repeated 3 times.
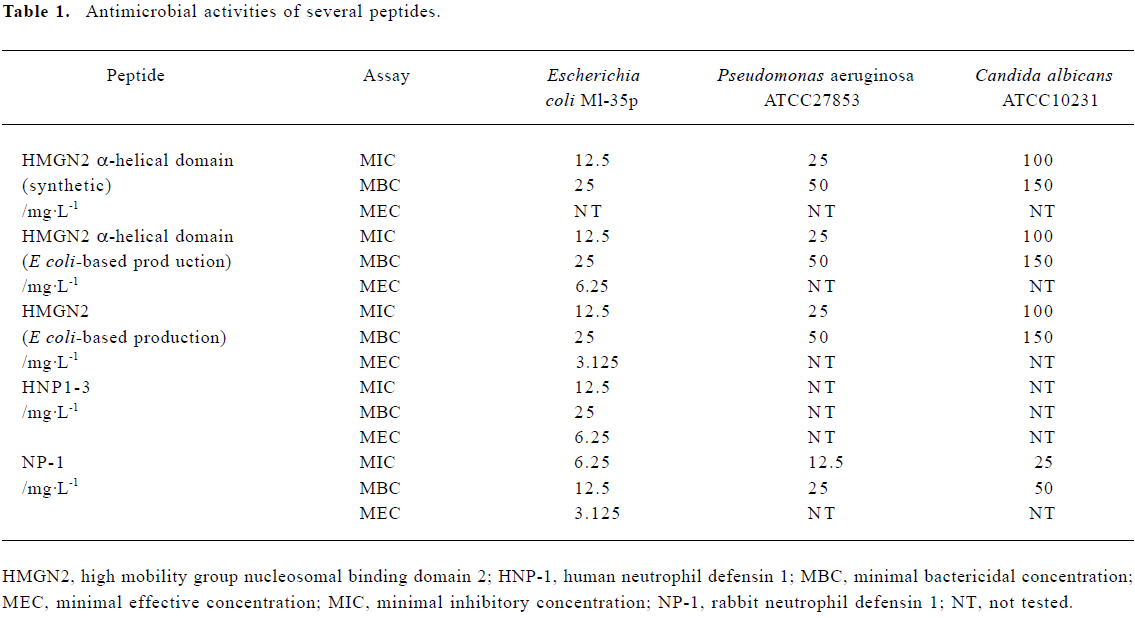
Full table
Discussion
High mobility group (HMG) proteins have been described as an abundant family of non-histone proteins in the cell nucleus of vertebrate and invertebrate organisms[15]. In the narrowest traditional sense, this HMG protein family consists of 6 proteins and is subdivided into 3 subfamilies: the HMG-box (HMGB) (formerly HMG-1/-2), the HMG AT-hook (HMGA) (formerly HMG-I/Y/C), and the HMG nucleosomal binding domain (HMGN) (formerly HMG-14/-17) subfamilies. Each of these classes seems to have a distinct function in the nucleus[16]. HMGN has a cell cycle-related cellular location[17]. The functional gene is located on chromosome 1p36.1[9], and it contains 6 exons, with an extremely high GC content and a “HTF” island, indicative of a housekeeping gene that could be crucial for regulating the function of cells[9]. How-ever, up to now, the biological role of this protein has not been clear. A variety of experiments have shown that HMGN2 is preferentially associated with chromatin subunits containing transcribed genes and enhances the transcriptional potential of corresponding genes[10,11]. Other experiments indicate that HMGN2 maintains the timing of early embryonic development in the mouse, and shows developmental regulation during organogenesis[12]. Furthermore, abnormal gene or protein expression of HMGN2 is related to some diseases such as neoplasms and autoimmune diseases[18,19]. The significance of HMGN2 in the host defense against infection is unclear. Frohm and colleagues attempted to identify antimicrobial polypeptides from human wound and blister fluid. Several known antimicrobial peptides or proteins, eg defensins HNP1–3, lysozyme, FALL-39 and histone H2B fragments, were found. Although HMGN2 was isolated, its antimicrobial property was not determined[20]. More recently, Fernandes et al have described a potent antimicrobial peptide isolated from the skin mucus secretions of fish[21], that is a member of the HMG protein family. In our experiment we observed the antimicrobial activity of the HMGN2 protein.
Many cationic antibiotic peptides are suggested to be membrane-active, assembling to form channels[22]. Alternatively, certain peptides cluster at the membrane surface cause a cooperative permeabilization by the carpet effect[23–25]. On the other hand, apidaecins function through a receptor-activated non-pore-forming mechanism involving stereospecificity[25]. PR-39 kills bacteria by interrupting both DNA and protein synthesis and the DNA binding property of tachyplesin I has also been implicated in antimicrobial activity[26]. By protein structure analysis of HMGN2, a transmembrane α-helical structure located at residues 18–48 was found, and this location is related to the DNA binding domain of this protein. As such, we prepared a recombinant α-helical domain and confirmed its antimicrobial activity. Thus, the transmembrane α-helical domain may be essential for the antimicrobial activity of HMGN2. The antimicrobial mechanisms of HMGN2 should be further studied.
References
- Boman HG. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol 1995;13:61-92.
- Garcia-Penarrubia P, Koster FT, Kelley RO, McDowell TD, Bankhurst AD. Antimicrobial activity of human natural killer cells. J Exp Med 1989;169:99-113.
- Levitz SM, Mathews HL, Murphy JW. Direct antimicrobial activity of T cell. Immunol Today 1995;16:387-90.
- Prokesova L, Dung DH, Trebichavsky I, Formankova E, Stepankova V, John C. Antimicrobial activity of human mononuclear leukocytes against Staphylococcus aureus. Folia Microbiol (Praha) 1994;39:428-34.
- Kumar J, Okada S, Clayberger C, Krensky AM. Granulysin: a novel antimicrobial. Expert Opin Investig Drugs 2001;10:321-9.
- Andersson M, Gunne H, Agerberth B, Boman A, Bergman T, Sillard R, et al. NK-lysin, a novel effector peptide of cytotoxic T and NK cells. Structure and cDNA cloning of the porcine form, induction by interleukin 2, antimicrobial and antitumor activity. EMBO J 1995;14:1615-25.
- Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, et al. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood 2000;96:3086-93.
- Yang H, Huang N, Wu Q, Wang B. Isolation and characterization of an antimicrobial polypeptide from human LAK cells. Chin J Microbial Immunol 2003;23:241-3.
- Landsman DO, McBride W, Bustin M. Chromosomal protein HMGN2. Complete human cDNA sequence and evidence for a multigene family. Nucleic Acids Res 1986;17:2301-14.
- Tremethick DJ, Hyman L. High mobility group protein 14 and 17 can prevent the close packing of nucleosomes by increasing the strength of protein contacts in the linker DNA. J Biol Chem 1996;271:12009-16.
- Vestner B, Bustin M, Gruss C. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMGN2. J Biol Chem 1998;273:9409-14.
- Lehtonen S, Lehtonen E. HMGN2 is an early marker of inductive interactions in the developing mouse kidney. Differentiation 2001;67:1541-63.
- Selsted ME, Szklarek D, Lehrer RI. Purification and antimicrobial activity of antimicrobial peptides of rabbit granulocytes. Infect Immun 1984;45:150-4.
- Ganz T, Selsted ME, Szklarek D, Harwig SSL, Daher K, Bainton DF, et al. Defensins, natural peptide antibiotics of human neotrophils. J Clin Invest 1985;76:1427-35.
- Postnikov YV, Herrera JE, Hock R, Scheer U, Bustin M. Cluster of nucleosomes containing chromosomal protein HMGN2 in chromatin. J Mol Biol 1997;274:454-65.
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility group chromosomal proteins. Mol Cell Biol 1999;19:5237-46.
- Hock R, Scheer U, Bustin M. Chromosomal protein HMG-14 and HMGN2 are released from mitotic chromosomes and imported into the nucleus by active transport. J Cell Biol 1998;143:1427-36.
- Spieker N, Beitsma M, Sluis PV, Roobeek I, Dunnen JTD, Speleman F, et al. An integrated 5-Mb physical, genetic, and radiation hybrid map of a 1p36.1 region implicated in neuroblastoma pathogenesis. Genes Chromosomes Cancer 2000;27:143-52.
- Ayer LM, Rubin RL, Dixon GH, Fritzler MJ. Antibodies to HMG proteins in patients with drug-induced autoimmunity. Arthritis Rheum 1994;37:98-103.
- Frohm M, Gunne H, Bergman AC, Agerberth B, Bergman T, Boman A, et al. Biochemical and antimicrobial analysis of human wound and blister fluid. Eur J Biochem 1996;237:86-92.
- Fernandes JMO, Saint N, Kemp GD, Smith VJ. Oncoryncin III: a potent antimicrobial peptide derived from the nonhistone chromosomal protein H6 of rainbow trout, Oncorhynchus mykiss. Biochem J 2003;373:621-8.
- Tiozzo E, Rocco G, Tossi A, Romeo D. Wide-spectrum antibiotic activity of synthetic amphipathic peptides. Biochem Biophys Res Commun 1998;249:202-6.
- Gennaro R, Zanetti M. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers 2000;55:31-49.
- Yang YX, Feng Y, Wang BY. PCR-based site-specific mutagenesis of peptide antibiotic FALL-39 and its biologic activities. Acta Pharmacol Sin 2004;25:239-45.
- Cabiaux V, Agerberth B, Johansson J, Homble F, Goormaghtigh E, Ruysschaert JM. Secondary structure and membrane interaction of PR-39, a Pro+Arg-rich antibacterial peptide. Eur J Biochem 1994;224:1019-27.
- Agerberth B, Lee JY, Bergman T, Carlquist M, Boman HG, Mutt V, et al. Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline–arginine-rich antimicrobial peptides. Eur J Biochem 1991;202:849-54.
