Exploring in vitro roles of siRNA in cardiovascular disease1
Introduction
Cardiovascular disease (CVD) is the leading cause of mortality and morbidity in the world. In recent years, the study of the molecular mechanisms of CVD has accelerated the discovery of many new potential therapeutic targets. Fire et al[1] injected double-stranded RNA (dsRNA) into the nematode worm Caenorhabditis elegans, resulting in silencing the gene whose sequence was complementary to that of the dsRNA. This post-transcriptional gene silencing (PTGS) phenomenon was named RNA interference (RNAi). Since then, it has become clear that RNAi is a defensive process of cells used to degrade invasive genetic materials and inhibit endogenous genes that should be controlled. This technology is an extremely useful tool for identifying gene functions and evaluating potential therapeutic targets. So, an increasing number of biotechnological and pharmaceutical companies are attempting to develop RNAi-based drugs for the prevention and treatment of human disease such as viral infections, tumors and CVD[2–4].
Many CVD are chronic in nature and worsen progressively over a long period of time. Despite the significant advances that have been made in the therapy of CVD, current available drugs are associated with many undesirable factors such as toxicity, complexity and resistance. As a natural self-defense mechanism of eukaryotic cells, RNAi can offer major advantages over pharmacological therapy by targeting pathogenic genes specific for CVD with high potency and low toxicity. RNAi has been used in models of hyperlipemia, ischemia-reperfusion, and choroidal neova-scularization and for identification of those genes involved in CVD[5,6]. This review will discuss some of the more subtle mechanistic aspects of RNA-induced PTGS, and present recent work that focuses on the potential application of this breakthrough technology to CVD such as congenital heart disease, hypertension, atherosclerosis, cardiac hypertrophy, myocarditis, and heart failure.
Molecular mechanisms
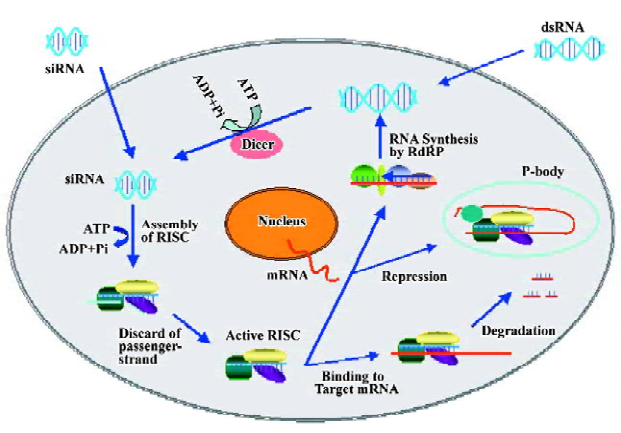
Biochemical and genetic studies have revealed the molecular mechanisms by which homologous dsRNA cause degradation of target messenger RNA. RNAi includes 2 main steps: an initiator step for generation of small-interfering RNAs (siRNAs) from long dsRNA or mature microRNAs (miRNA) from their precursors, and an effector’s step for cleavage or repression of target RNA[7].
In the initiator step, the dsRNA-specific endonuclease Dicer binds with high affinity to dsRNA of more than 38 bp in length and chops long dsRNA (introduced directly or via a transgene or virus) into fragments of ~22 nt[8]. The primary structure of Dicer includes an ATP-dependent RNA helicase domain, a Piwi/Argonaute/Zwille (PAZ) domain, two RNase III-like domains and a COOH-terminal dsRNA-binding domain. The recently identified Dicer 1 and Dicer 2 are responsible for the production of mature miRNAs from their precursors and for the cleavage of long dsRNA into siRNAs, respectively[9]. siRNA is composed of 21–23 nt dsRNA duplexes with a 5'-monophosphate, a 3'-hydroxyl group and 2-nt 3' overhangs, and this configuration plays an important role in the efficacy of silencing.
In the effector’s step, the siRNA duplexes are incorporated into the RNA-induced silencing complex (RISC), a protein-RNA effector nuclease complex that recognizes and destroys passenger-strand of siRNA duplexes[10]. After releasing that cleaved strand, the guide-strand in active RISC becomes available to interact with target mRNA. The phosphorylation of siRNA 5'-terminal is required to entry into RISC. The PAZ and PIWI domains of RISC can recognize the 3'-terminus and the 5'-terminus of the guide strand, respectively, and then targets the homologous transcript by base-pairing interactions with the guide strand. Finally, an RNase H region at the PIWI domain cleaves the mRNA between the tenth and eleventh nucleotide from the 5' terminus of the siRNA[11,12].
One of the most important characters of RNAi is the ability to amplify and sustain gene silencing induced by dsRNA in many organisms, even when triggered by minute quantities of aberrant RNA. Recently genetic studies have found that siRNA might act as mRNA-specific primers that are incorporated during the subsequent conversion of the target mRNA into dsRNA. Nascent dsRNA is then cleaved by RNase III-related enzymes to degrade the mRNA while generating new siRNAs in the process. RNA-directed RNA polymerase (RdRP) plays the crucial role in mediating the incorporation of a synthetic siRNA into nascent dsRNA. This enzyme is discovered in most eukaryote except mammals and insects[13]. It is not clear if the cardiac myocytes connected by gap-junctions contain RdRP and display RNAi spread between cells. Through this approach, aberrant RNAs can be degraded efficiently through a cycle of “degradative-PCR” (Figure 1).
Advantages
In comparison with other conventional drugs, siRNA have many advantages: (1) The selection of target sites is much easier and more flexible because target mRNA and siRNA are sequences-specific and complementary. For a given mRNA molecule, the inhibitory effects of siRNA can be achieved by targeting different regions of target mRNA[4]; (2) For gene silencing, only a substoichiometric amount of siRNA is enough to decrease homologous mRNA drastically within 24 h; (3) siRNA can knockdown the expression of any cognate genes in cells of different species even though that gene is critical for animal development; (4) siRNA do not seem to adversely affect the physiological functions of the cells. The given length and high level of homology of siRNA to the target region of cognate transcription ensure the selective destruction of only the transcript of interest. siRNA without suitable targets seems to remain inert within cells. This exclusive specificity without adverse side effects is the most attractive feature of using RNAi for an antiviral approach[6,7]; (5) siRNAs can silence genes stably. With the application of plasmid vectors and viral vectors, siRNA can display their long-term biological effects[14]. Taken together, siRNAs produced in vivo or in vitro transfected into cultured cells or animals could result in the sequence-specific silencing of mRNA. With the proof-of-concept studies, siRNA-based gene drugs will be used as an alternative therapeutic strategy in the future.
Disadvantages and improvements
Although siRNA-mediated RNAi has become a functional genomic tool, recent studies in vitro have revealed that some duplex siRNA sequences have off-target effects, trigger an interferon response[15] and possess other drawbacks such as higher cost, short duration, and hard delivery into specific tissues and resistances. Therefore, many new methods are being developed to overcome these disadvan-tages.
Researchers have proposed general guidelines for designing siRNA oligonucleotides[16]. Recently, Heale et al proposed a novel approach for the determination of mRNA secondary structures and showed, that in combination with duplex-end energies, the predicted strong secondary structures could account for 80% of non-functional siRNA target sites[17]. Bioinformatics have also been used to design siRNA, and many websites provide methods to pick siRNAs. Our laboratory has applied for a patent to design siRNA, and the success rate of gene silencing is more than 90%.
In order to improve the efficiency and duration of small RNA in gene silencing research, several methods to produce siRNAs have been proposed, such as chemically synthesized siRNA[18], DNA vector-based siRNA, and siRNA cassette[19]. The chemical modifications of siRNA has been shown to be necessary for increasing the efficiency of small RNAs such as the stability of siRNAs within the body, bioavailability to different tissues, affinity for the blood proteins, and specific delivery to the chosen site[20]. Chemically stabilized siRNAs with partial phosphorothioate backbone and 2'-O-methyl sugar modifications on the sense and the antisense strands showed significantly enhanced resistance towards degradation by exonucleases and endonucleases in serum and in tissue homogenates. Taken together, the chemical modifications of siRNAs may improve the stability and utility of siRNAs for therapeutic application in vivo.
Delivery system
Recent rapid progress in the application of RNAi to mammalian cells offers new approaches to drug target identification and validation; however, the use of RNAi in mammals has been hindered by the inability to deliver siRNAs effectively. To make RNAi highly efficient in cardiomyo-cytes, vascular smooth muscle cells (VSMC), and vascular endothelial cells with low transfection efficacy, virus-mediated RNAi has been developed[18,19]. The effects of RNAi on GAPDH transcripts were reduced by 90% in the primary cultured cells, indicating that virus-mediated gene silencing is a promising technique for gene suppression in cardiovascular studies[21].
As an emerging technique with potential use as a therapy for CVD, ultrasound-targeted microbubble destruction (UTMD) has proven to be an efficacious method for delivering genes such as decoy oligodeoxynucleotides and anti-sense oligonucleotide for TNF-α[22,23]. For the first time, Shohet et al found that UTMD could deliver recombinant adenovirus containing β-galactosidase to the heart, achieving transient transgene expression with striking tissue specificity[24]. Recently, UTMD has been used to deliver gene to cure ischemic heart diseases[25,26]. It appears that this new technique can be used for the delivery of siRNA to take advantage of its favorable characteristics such as organ specificity, low level of toxicity, no immunogenicity, repeatable applicability, and low costs.
Injection, a novel therapeutic method of siRNA delivery, includes local and systemic injection. Chae et al found that local injection of siRNA targeting S1P, a family of S1P G protein-coupled receptors required for the stabilization of nascent blood vessels during embryonic development, could silence the cognate transcript in endothelial cells and inhibited endothelial cell migration[27]. As a kind of systemic injection, hydrodynamic injection involves the rapid injection of a large-volume bolus to cause transient high venous pressures, which can facilitate the delivery of siRNAs. Hamar et al used hydrodynamic transfection to administer siRNAs targeting Fas into mice after ischemic reperfusion injury and demonstrated a 4-fold reduction of Fas mRNA and protein expression, and a substantial reduction of renal tubular apoptosis[28].
More interestingly, Soutschek et al developed a new delivery system by conjugation of cholesterol to siRNAs. They found that Chol-apoB-siRNAs, but not unconjugated apoB-siRNAs, could effectively degrade apolipoprotein B mRNA by more than 50% in the liver and by 70% in the jejunum via injection into a tail vein[6]. The plasma levels of the apoB protein were reduced also by more than 60%. Furthermore, chol-apoB-siRNA resulted in about a 40% reduction in the levels of LDL and total plasma cholesterol. Soutschek’s work is of significance for the systemic in vivo application of RNAi technology, because it shows for the first time a new class of therapeutics that harnesses the RNAi mechanism and suggests the therapeutic potential of RNAi for the treatment of CVD. Recently, it has been reported that apoB-specific siRNAs were encapsulated in stable nucleic acid lipid particles and administered by intravenous injection to cynomolgus monkeys. The results demonstrated that the apoB protein, serum cholesterol and low-density lipoprotein levels significantly decreased as early as 24 h after treatment for 11 d[5]. Although these delivery systems are effective in animal models, they still need detailed safty testing and efficacy evaluation before they are used for human clinical trials.
RNAi application in CVD
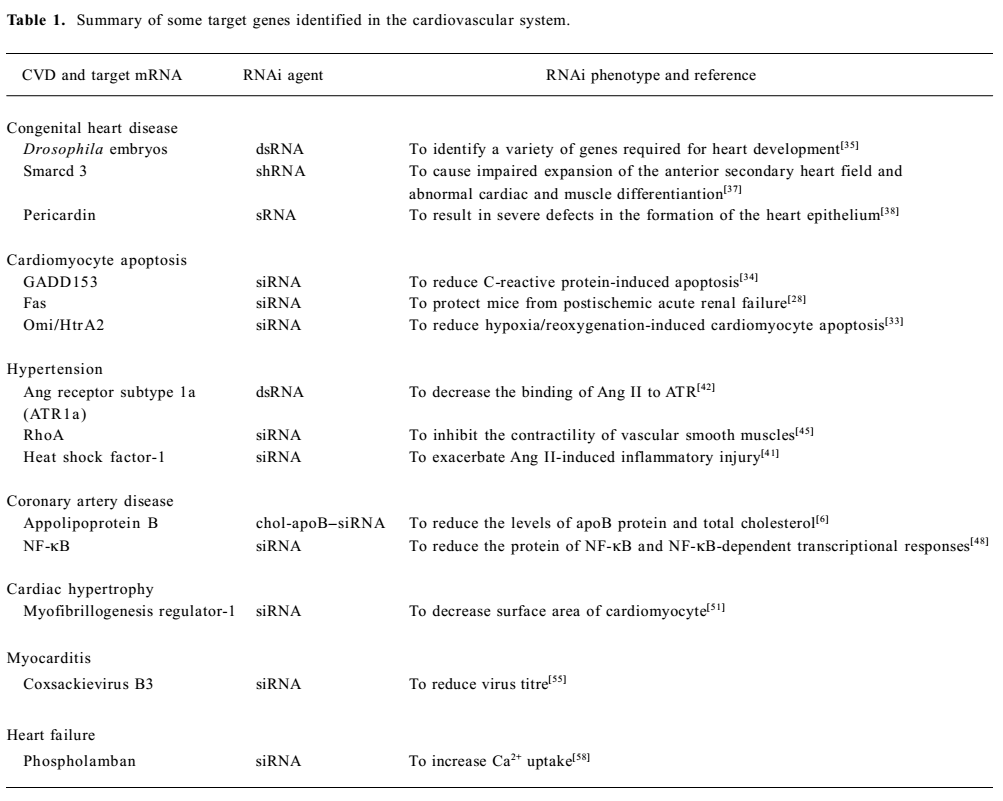
It has been established that the activity of a gene can be inhibited by the introduction of dsRNA with the sequence specific to the gene. The specificity and potency of RNAi make it ideal for investigating human disease-susceptibility genes[29]. In this way, some CVD-specific molecular targets have been identified (see Table 1 for a summary). So RNAi-based gene drugs can be developed in the near future.
Full table
Apoptosis in CVD
Compelling evidence has accumulated indicating that apoptosis may play a critical role in the pathogenesis of a variety of CVD including atherosclerosis, myocardial infarction, ischemia followed by reperfusion, and heart failure. Naturally, there is intensive apoptosis research in the field of CVD therapy.
Evidence has confirmed that Fas-induced apoptosis can enlarge infarct size during reperfusion of ischemic tissue in multiple tissues such as the heart[30], kidney and brain. Hamar et al found that siRNAs targeting Fas could inhibit Fas expression in the murine kidney in vivo and protect mice from postischemic acute renal failure[28]. Another good example is that in vivo silencing the Fas gene protects mice from liver failure and fibrosis by intravenous injection of Fas siRNA. Therefore, these findings suggest that the heart or brain might also be protected from ischemia reperfusion injury by silencing Fas, and offer a novel therapeutic that is prophylactic against ischemia-perfusion disease. On the other hand, experimental results show that excessive apopto-sis of vascular smooth muscle cells (VSMCs) plays a key role in the progression of atherosclerotic lesions, resulting in many cardiovascular events[32]. GADD153, a member of the CCAAT/enhancer-binding protein (C/EBP) family of transcription factors, has been linked to apoptosis in VSMCs. Blaschke et al found that inhibition of GADD153 by siRNAs reduces C-reactive protein-induced GADD153 mRNA expression and apoptosis. Moreover, several recent investigations have demonstrated that Omi/HtrA2, a serine protease, could be released into the cytosol from mitochondria and promote caspase-dependent apoptosis after an apoptotic insult. Liu et al found that the introduction of siRNA molecules against Omi into cardiomyocytes effectively eliminated Omi/HtrA2 protein expression and reduced hypoxia reoxygenation-induced cardiomyocyte apoptosis with decreased caspase-3 activity[33]. Their findings might lead to a new strategy for the treatment of cardiovascular disorders[34].
Congenital heart disease
Identifying genetic components plays a key role in understanding development processes that can be useful for the discovery of targets in the treatment of CVD. Some laboratories have utilized the technology of RNAi to identify genes involved in heart development. With the use of an RNAi-based genome wide loss-of-function screen, Kim’s group identified a variety of genes encoding cell-signaling proteins and transcription factors for different steps during the development of the Drosophila embryonic heart[35]. Recently, Qian et al revealed that neuromancer (nmr), a homolog of Tbx20, is another determinant of morphogenesis of the Drosophila heart by conducting RNAi experiments with related genes. For example, reducing nmr function in the absence of pannier further aggravates the deficit in cardiac mesoderm specification[36]. Furthermore, another research group found that RNAi-mediated depletion of Smarcd 3, a 60-kDa subunit of the BAF complexes, in mouse embryos derived from embryonic stem cells caused defects in heart morphogenesis. The defect displays an impaired expansion of the anterior/secondary heart field that can lead to abnormal cardiac muscle differentiation[37]. More interes-tingly, Chartier et al reported that embryos in which pericardin, an extracellular matrix component, silenced via RNAi, could exhibit severe defects in the formation of the heart epithelium because pericardin can mediate the crosstalk between the dorsal ectoderm and cardioblasts required to regulate their coordinated movement during dorsal closure. In addition, they found that the heart epithelium displayed a disorganized appearance during its migration to the dorsal midline in these embryos following the knockdown of pericardin mRNA[38]. Obviously, the silencing of pericardin is associated with pathologic holes in the walls of the heart. Taken together, these studies demonstrate that RNAi is an efficient reverse genetic tool for producing and identifying the loss-of-function mutant phenotypes of genes involved in CVD.
Hypertension
Hypertension is one of the most important risk factors for stroke, congestive heart failure, myocardial infarction, and peripheral vascular disease. Several lines of experimental results have revealed that renin-angiotension systems (RAS) are involved in the development and maintenance of hypertension. Angiotensin (Ang) II is a potent vasoconstrictor peptide produced by the RAS, and binds to 2 distinct receptor subtypes, namely type 1 (AT1) and type 2 (AT2). Mice with a homozygous deletion of the AT1A subtype (AT1A–/–) exhibit reduced blood pressures without pressor responsible for Ang II infusion[41]. It implicates AT1A as the primary subtype accountable for Ang II actions in mice. Although several AT1 receptor antagonists are used for the treatment of hypertension, they pose significant limitations. So, it is important to develop new ways to improve hypertension control by providing longer-lasting effects with a single dose and reducing side effects that lead to poor com-pliance. Recently, Vazquez et al employed RNAi technology to explore these possibilities[42]. They selected AT1R as the target gene to design corresponding dsRNAs, and then transfected them into Chinese hamster ovary cells (CHO), which express rat AT1R. They found that transfection of AT1R-expressing CHO cells with AT1R-dsRNA resulted in an 80% decrease in the level of AT1R mRNA. Furthermore, dsRNA caused a dose-dependent decrease in the specific binding of Ang II to AT1R-expressing CHO cells. To determine whether the decrease in AT1R -specific binding was associated with a reduction in functional AT1R, they examined the effects of dsRNA transfection on Ang II-stimulated calcium uptake. The Ang II-induced increase in calcium uptake was completely abolished in AT1-dsRNA transfected cells. On the other hand, Chen et al found that knocking down the expression of heat shock factor-1 (HSF-1) with RNAi technology exacerbated Ang II-induced inflammatory injury by causing significantly higher activation of NF-κB in VSMCs[43]. Such evidence would support the notion that heat shock proteins play a direct role in suppressing Ang II-induced inflammatory signaling pathways and subsequent inflammation. Furthermore, recent research on the mechanism of hypertension showed that the RhoA-Rho kinase pathway was the important pathogenesis of the abnormal contraction of the VSMCs in CVD[44]. Bi et al discovered that the knockdown of RhoA by RNAi decreased the level of RhoA mRNA and the contractility of the cultured VSMCs[45]. Their result indicated that the expression level of RhoA played a critical role in the regulation of contractility in the de-differentiated VSMC, and RhoA could be a new therapy target of hypertension. Taken together, these findings suggest that RNAi might have potential as an alternative to drug therapy for hypertension.
Atherosclerosis
Atherosclerosis is a chronic inflammatory disease of the arterial intima, resulting from a concerted action of multiple factors[46]. Many studies have shown that macrophages and T-cells play critical roles in multiple aspects of the pathogenesis of the disease. Further molecular analysis indicates that the nuclear factor-kappa B (NF-κB) plays a prominent role in the formation of atherosclerosis because of its ability to adhere to elements in the promoters of key inflammatory and atherosclerosis genes[47]. Dwarakanath et al synthesized siRNA targeting NF-κB by the use of a rapid PCR-based approach that generates sense and antisense siRNA separated by a hairpin loop downstream of the U6 promoter, and then transfected them into the VSMC derived from 12/15-Loko mice versus genetic control wild type mice in relation to cellular growth and migration[48]. They found that the mRNA and protein of NF-κB and NF-κB-dependent transcriptional responses were reduced markedly by the siRNA. On the other hand, Kobashi et al found that adiponectin enhances Akt phosphorylation[49]. The inhibitory effect of adiponectin on TNF-α-induced interleukin (IL)-8 synthesis was blocked in part by pretreatment with the PI3 kinase inhibitor LY294002 or by Akt siRNA transfection. It suggests that Akt activation might inhibit IL-8 synthesis, a pro-inflammatory chemokine that plays a role in atherogenesis. These observations may suggest new options for the treatment of atherosclerosis.
Cardiac hypertrophy
Cardiac hypertrophy is a compensatory response to a variety of physiological or pathological stimuli, and prolonged hypertrophic responses may eventually lead to arrhy-thmia, heart failure and sudden death[50]. So identifying the cardiac hypertrophy-related novel human genes will provide important insights into the mechanisms that regulate hypertrophic cell growth and assist in development of new pathway for treatment of cardiac hypertrophy heart failure.
Liu et al identified a novel human gene, myofibrillogenesis regulator-1 (MR-1) from a human skeletal muscle cDNA library that interacts with contractile proteins and exists in human myocardial myofibrils[51]. They established a hypertrophy model in which hypertrophic cell growth can be induced by Ang II incubation in cultured neonatal rat cardio-myocytes. By transfecting neonatal cardiomyocytes with a pSi-1 targeting the MR-1 sequence, Liu found that the MR-1 mRNA and protein expression were greatly decreased. Furthermore, compared with the Ang II-treated group, the MR-1 RNAi+Ang II group showed a decrease on the surface area of cells by 36%. More interestingly, Pedram et al[52] reported that the translocation of the hypertrophic transcription factor, NF-AT, to the nucleus of the cardiomyocyte and the enhancement of NF-AT transcriptional activity induced by Ang II could be prevented by 17β-estradiol (E2). Ang II also stimulated the activation of ERK and protein kinase C, contributing to cardiac hypertrophy. E2 inhibited these pathways, related to the stimulation of atrial natriuretic peptide production and secretion. These observations were further supported by the evidence that siRNA against the MCIP1 gene significantly reversed both the E2 restraint of protein synthesis and the inhibition of Ang II-induced calcineurin activity. Accordingly, their findings may provide a better understanding of the mechanism of cardiac remodeling and new insight into the development of novel therapeutic strategies in cardiac hypertrophy.
Myocarditis
Evidence has accumulated that viral myocarditis is an important cause of heart failure and dilated cardiomyopathy[53]. More effective approaches are needed to treat viral infection. If genes responsible for affective disorders are identified, gene silencing could be an alternative therapeutic tool, especially for cases of drug therapy resistance.
Coxsackievirus B3 (CVB3) has been identified as the most common causal agent of viral myocarditis, but existing drug therapies are of limited value[54]. Many studies have shown that RNAi can control viral infection by targeting viral genes. Recently, Schubert et al found that 2 independent siRNA targeting the 3D RNA-dependent RNA polymerase were able to reduce virus titre by 80% and 90%, respectively[55]. Their results demonstrate the enormous potential of the RNAi approach. More interestingly, Schubert and colleagues constructed a siRNA double expression vector (SiDEx) to achieve simultaneous expression of both siRNA from 1 plasmid. Compared with conventional expression vectors, SiDEx showed substantial gene regulation of the mutated target RNA. So it is believed that SiDEx may be a helpful tool to achieve sustained silencing of viruses, ultimately reducing the risk of emergence of viable mutants. Recently, Yan et al reported that the siRNA targeting the viral protease 2A displayed a 92% inhibition of CVB3 replication and a 90% protection of the siRNA-pretreated cells. Moreover, they found that administration of the siRNA after viral infection could effectively inhibit CVB3 replication, indicating its therapeutic potential[56]. These findings imply that siRNA-based gene drugs may be an effective therapy for viral myocarditis.
Heart failure
Investigation shows that heart failure is a common lethal condition associated with various CVD and remains the leading cause of morbidity and mortality. One of the important features of heart failure is a decreased Ca2+ uptake into the sarcoplasmic reticulum (SR) by the SR Ca2+-ATPase 2 (SERCA2), which is negatively regulated by phospholamban (PL), a key regulator of cardiac calcium homeostasis[57].
Recent findings demonstrated that the development of severe heart failure in the genetic MLP (-/-) animal model could be abolished completely by the targeted ablation of PL. PL has been considered as the potential therapeutic target for the improvement of SR Ca2+ uptake and cardiac function. Watanabe et al synthesized 21 nt siRNA duplexes with symmetric 2-nt 3'-overhangs targeting PLB mRNA and introduced them into neonatal rat cardiac myocytes by use of the HVJ envelope vector[58]. They found that PLB siRNA resulted in a significant decrease in the levels of both PLB mRNA and protein, while the mRNA and protein of SERCA2 calsequestrin were not affected. The affinity of SERCA2 for Ca2+ was also increased. In order to determine the effect of PLB RNAi on the cardiac myocytes in which Ca2+ handling was impaired, Watanabe et al exposed myocytes to H2O2, a reactive oxygen species. The same result with decreased PLB mRNA and protein levels was achieved. So Watanabe’s strategy used for the PL ablation may be considered as a novel and attractive candidate for clinical therapy in heart failure.
Conclusion
CVD is a severe public health problem with significant personal, social, and economical consequences. More effective approaches are urgently needed to treat CVD. It is evident from the above discussion that the use of RNAi may hold great promise for a permanent treatment of CVD by targeting special pathogenic genes. Given the relative ease with which one can design one to several siRNA to target a gene of interest, there is no need for time-consuming pharmacological analyses and drug specificity studies. One can simply target the specific genes in the cardiovascular system by using chemically synthesized siRNA or viral vectors expressing hsRNA. The potential targets in CVD are numerous. However, careful assessment is required for the potential of RNAi as a gene therapy approach for controlling CVD, because there are some questions to be solved such as the delivery of vector and dosage required to elicit the desired effect. In addition, because each gene may be a point of signaling networks, and cross talk with other signal pathways, inhibition of the signaling certainly affects the intricate networks and may leads to unwanted side effects. So it has no exaggeration to say that careful planning is necessary before any clinical trial, although a report has shown an encouraging result that a siRNA targeting vascular endothelial growth factor receptor-1 has potential for clinical use in preventing aging-related macular degeneration.
Even although RNAi-based therapy is still in infancy stage, it holds tremendous promise for use in routine clinical practice as an adjunct to existing procedures since it can help to overcome the limitations associated with current therapeutic regimen[59,60]. The increasing knowledge of small RNA will help us to better understand the mechanism of gene expression regulation in eukaryotes. Therefore, modulation of transcripts of interest may be an alternative molecular strategy for experimental research and treatment of pathophysiological states[61,62]. Scientists have developed many RNAi libraries to study the function of genes in nematodes. With the advent of an RNAi library in mammals and the refinement of techniques to silence gene, siRNA-based drugs will make great advances in the prevention and treatment of CVD.
References
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806-11.
- de Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature 2005;436:953-60.
- Yin JQ, Gao J, Shao R, Tian WN, Wang J, Wan Y. siRNA agents inhibit oncogene expression and attenuate human tumor cell growth. J Exp Ther Oncol 2003;3:1-11.
- Drosett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 2004;3:319-29.
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature 2006;441:111-4.
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004;432:173-8.
- Hannon GJ. RNA interference. Nature 2002;418:244-51.
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000;404:293-6.
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 2004;11:214-8.
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005;123:607-20.
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004;305:1437-41.
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005;434:666-70.
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science 2005;309:1519-24.
- Paddison PJ, Hannon GJ. siRNAs and shRNAs: skeleton keys to the human genome. Curr Opin Mol Ther 2003;5:217-24.
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA 2004;101:1892-7.
- Santoyo J, Vaquerizas JM, Dopazo J. Highly specific and accurate selection of siRNAs for high-throughout functional assays. Bioinformatics 2005;21:1376-82.
- Heale BS, Soifer HS, Bowers C, Rossi JJ. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res 2005;33:e30.
- lmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, . Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 2005; 33: 439–47.
- Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett 2005;579:5974-81.
- Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol 2004;8:570-9.
- Kasahara H, Aoki H. Gene silencing using adenoviral RNAi vector in vascular smooth muscle cells and cardiomyocytes. Methods Mol Med 2005;112:155-72.
- Hashiya N, Aoki M, Tachibana K, Taniyama Y, Yamasaki K, Hiraoka K, et al. Local delivery of E2F decoy oligodeoxynucleo-tides using ultrasound with microbubble agent (Optison) inhibits intimal hyperplasia after balloon injury in rat carotid artery model. Biochem Biophys Res Commun 2004;317:508-14.
- Erikson JM, Freeman GL, Chandrasekar B. Ultrasound-targeted antisense oligonucleotide attenuates ischemia/reperfusion-induced myocardial tumor necrosis factor-alpha. J Mol Cell Cardiol 2003;35:119-30.
- Shohet RV, Chen S, Zhou YT, Wang Z, Meidell RS, Unger RH, et al. Echocardiographic destruction of albumin microbubbles directs gene delivery to the myocardium. Circulation 2000;101:2554-6.
- Wang ZG, Liang ZY, Ran HT, Ren H, Zhang QX, Huang AL, et al. Ultrasound-mediated microbubble destruction enhances VEGF gene delivery to the infarcted myocardium in rats. Clin Imaging 2004;28:395-8.
- Kondo I, Ohmori K, Oshita A, Takeuchi H, Fuke S, Shinomiya K, et al. Treatment of acute myocardial infarction by hepatocyte growth factor gene transfer: the first demonstration of myocardial transfer of a “functional” gene using ultrasonic microbubble destruction. J Am Coll Cardiol 2004;44:644-53.
- Chae SS, Paik JH, Furneaux H, Hla T. Requirement for sphingosine 1–phosphate receptor-1 in tumor angiogenesis demonstrated by in vivo RNA interference. J Clin Invest 2004;114:1082-9.
- Hamar P, Song E, Kokeny G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci USA 2004;101:14883-8.
- Yin JQ, Wan Y. siRNA-mediated gene regulation system. Int J Mol Med 2002;10:355-65.
- Li Y, Takemura G, Kosai K, Takahashi T, Okada H, Miyata S, et al. Critical roles for the Fas/Fas ligand system in postinfarction ventricular remodeling and heart failure. Circ Res 2004;17:627-36.
- Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003;9:347-51.
- Bennett MR. Apoptosis of vascular smooth muscle cells in vascular remodelling and atherosclerotic plaque rupture. Cardiovasc Res 1999;41:361-8.
- Liu HR, Gao E, Hu A, Tao L, Qu Y, Most P, et al. Role of Omi/HtrA2 in apoptotic cell death after myocardial ischemia and reperfusion. Circulation 2005;111:90-6.
- Blaschke F, Bruemmer D, Yin F, Takata Y, Wang W, Fishbein MC, et al. C-reactive protein induces apoptosis in human coronary vascular smooth muscle cells. Circulation 2004;110:579-87.
- Kim YO, Park SJ, Balaban RS, Nirenberg M, Kim Y. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc Natl Acad Sci USA 2004;101:159-64.
- Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev Biol 2005;279:509-24.
- Lickert H, Takeuchi JK, Von BI, Walls JR, McAuliffe F, Adamson SL, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 2004;432:107-12.
- Chartier A, Zaffran S, Astier M, Semeriva M, Gratecos D. Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development 2002;129:3241-53.
- Kizer JR, Wachtell K, Lehto M, Julius S, Beevers G, de Faire U, et al. Stroke reduction in hypertensive adults with cardiac hypertrophy randomized to losartan versus atenolol: the losartan intervention for endpoint reduction in hypertension study. Hypertension 2005;45:46-52.
- Watanable T, Barker TA, Berk BC. Angiotensin II and the endothelium: diverse signals and effects. Hypertension 2005;45:163-9.
- Gurley SB, Le TH, Coffman TM. Gene-targeting studies of the renin-angiotensin system: mechanisms of hypertension and cardiovascular disease. Cold Spring Harb Symp Quant Biol 2002;67:451-7.
- Vazquez J, Correa de Adjounian MF, Sumners C, Sumners C, Gonzalez A, Diez-Freire C, et al. Selective silencing of angiotensin receptor subtype 1a (AT1aR) by RNA interference. Hypertension 2005;45:115-9.
- Chen Y, Currie RW. Small interfering RNA knocks down heat shock factor-1 (HSF-1) and exacerbates pro-inflammatory activation of NF-kappaB and AP-1 in vascular smooth muscle cells. Cardiovasc Res 2006;69:66-75.
- Sward K, Mita M, Wilson DP, Deng JT, Susnjar M, Walsh MP. The role of RhoA and Rho-associated kinase in vascular smooth muscle contraction. Curr Hypertens Rep 2003;5:66-72.
- Bi D, Nishimura J, Niiro N, Hirano K, Kanaide H. Contractile properties of the cultured vascular smooth muscle cells. The crucial role played by RhoA in the regulation of contractility. Circ Res 2005;96:890-7.
- Hansson SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2002;404:293-6.
- Collins T, Cybulsky MI. NF-kappaB: pivotal mediator or innocent bystander in atherogenesis? J Clin Invest 2001;101:255-64.
- Dwarakanath RS, Sahar S, Reddy MA, Castanotto D, Rossi JJ, Natarajan R. Regulation of monocyte chemoattractant protein-1 by the oxidized lipid, 13-hydroperoxyoctadecadienoic acid, in vascular smooth muscle cells via nuclear factor-kappa B (NF-kappaB). J Mol Cell Cardiol 2004;36:585-95.
- Kobashi C, Urakaze M, Kishida M, Kibayashi E, Kobayashi H, Kihara S, et al. Adiponectin inhibits endothelial synthesis of interleukin-8. Circ Res 2005;97:1216-9.
- Liu X, Li T, Sun S, Yang Y. Role of myofibrillogenesis regulator-1 in myocardial hypertrophy. Am J Physiol Heart Circ Physiol 2005;290:H279-85.
- Si X, Rahmarni M, Yuan J, Lou H. Detection of cardiac signaling in the injured and hypertrophied heart. Methods Mol Med 2005;112:291-303.
- Pedram A, Razandi M, Aitkenhead M, Levin ER. Estrogen inhibits cardiomyocyte hypertrophy : Antagonism of calcineurin- related hypertrophy through induction of MCIP1. J Biol Chem 2005; 280: 26 339–48.
- Liu P, Martino T, Opavsky MA, Penninger J. Viral myocarditis: balance between viral infection and immune response. Can J Cardiol 1996;12:935-43.
- Yang D, Yu J, Luo Z, Carthy CM, Wilson JE, Liu Z, et al. Viral myocarditis identification of five differentially expressed genes in coxsackievirus B3–infected mouse heart. Circ Res 1999;84:704-12.
- Schubert S, Grunert HP, Zeichhardt H, Werk D, Erdmann VA, Kurreck J. Maintaining inhibition: siRNA double expression vectors against coxsackieviral RNAs. J Mol Biol 2005;346:457-65.
- Yuan J, Cheung PK, Zhang HM, Chau D, Yang D. Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. J Virol 2005;79:2151-9.
- Hobai IA, O’Rourke B. Decreased sarcoplasmic reticulum calcium content is responsible for defective excitation-contraction coupling in canine heart failure. Circulation 2003;103:1577-84.
- Watanabe A, Arai M, Yamazaki M, Koitabashi N, Wuytack F, Kurabayashi M. Phospholamban ablation by RNA interference increase Ca2+ uptake into rat cardiac myocyte sarcoplasmic reticulum. J Mol Cell Cardiol 2004;37:691-8.
- Liu TG, Yin Q, Shang BY, Min Z, He HW, Jiang JM, et al. Silencing of hdm2 oncogene by siRNA inhibits p53-dependent human breast cancer. Cancer Gene Ther 2004;11:748-56.
- Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J Biotechnol 2006;124:12-5.
- Lu PY, Xie F, Woodle MC. In vivo application of RNA inter-ference: from functional genomics to therapeutics. Adv Genet 2005;54:117-42.
- Barik S. Development of gene-specific double-stranded RNA drugs. Ann Med 2004;36:540-51.
