Silencing livin gene by siRNA leads to apoptosis induction, cell cycle arrest, and proliferation inhibition in malignant melanoma LiBr cells
Introduction
Livin, also called melanoma inhibitor of apoptosis proteins (IAP)or kidney IAP, is a recently discovered inhibitor member of the apoptosis protein (IAP) family[1–3]. Similar to other IAP that are able to block apoptosis in a caspase-dependent or -independent manner, livin is selectively expressed in most human neoplasms, but not or to a lesser extent in normal differentiated tissues, and appears to be involved in tumor cell resistance to apoptosis induced by a variety of stimuli[4]. Moreover, livin has a differential expression pattern and higher expression rate in tissue samples and primary cultures derived from malignant melanoma patients, as well as in malignant melanoma cell lines[1,2,5,6]. In addition, high levels of livin expression were correlated with tumor progression and a lower survival rate, as well as the resistance of the cells to chemotherapy both in vitro and in melanoma patients receiving chemotherapy[5]. Therefore, the overexpression of livin renders malignant melanoma cells resistant to apoptotic stimuli and potentially contributes to the pathogenesis of this malignancy.
The discovery that small interfering RNA (siRNA) duplexes can trigger RNA interference (RNAi) for post-transcriptional gene silencing to knock down the expression of target genes in mammalian cells has opened the innovative access to developing the technique into therapeutics[7]. Over the past few years, RNAi-based therapies have been successfully implemented in a variety of cancer models[8]. The overexpression of livin, especially in malignant melanoma cells, implies that livin may be a potential target of RNAi by knocking down its expression to modulate the apoptosis deficiency for gene therapy in malignant melanoma.
In this study, to investigate the biological effect of silencing the livin gene on human malignant melanoma LiBr[9] cells, three chemically synthesized siRNAs targeting to livin were transiently transfected into LiBr cells and the effects on apoptosis, cell cycle and proliferation were observed in vitro.
Materials and methods
Cell line and reagents Malignant melanoma cell line LiBr[9] was a kind gift from Professor Tian-wen GAO (Fourth Military Medical University, Xi’an, China). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), Lipofectamine 2000, and TRIzol were purchased from Invitrogen (Carlsbad, CA, USA). The SYBR ExScript RT-PCR kits were from TaKaRa (Dalian, China). The mouse monoclonal antibody to livin (human; 88C570) was from Alexis Biochemicals (Lausen, Switzerland). The rabbit polyclonal antibody to caspase-3 (human; sc-7148) was from Santa Cruz (Santa Cruz, CA, USA). The rabbit polyclonal antibody to β-actin (human) was from Zhongshan (Beijing, China). Goat anti-mouse immunoglobulin G (IgG)/horseradish peroxidase (HRP) and goat anti-rabbit IgG/HRP were also from Zhongshan (China). The in situ cell death assay kit was from Keygen (Nanjing, China). The Annexin V-fluorescein-isothiocyanate (FITC) kit was from Jingmei Biotech (Shenzhen, China).
siRNA design and preparation The siRNA targetting to livin was designed according to the characterization of siRNA by Elbashir et al[7] and Reynolds et al[10]. As livin has 2 splicing variants, livin α and livin β[11], and the 2 isoforms possess an identical 843 bp sequence, except for an additional 54 bp in livin α, all 3 designed siRNA target livin β (GenBank Accession N
Cell culture and transfection The LiBr cells were cultured in DMEM supplemented with 10% FBS and without antibiotics at 37 ℃ in a humidified 5% (v/v) CO2 incubator. Transfection with siRNA was carried out with Lipofectamine 2000 according to the procedure recommended by the manufacturer.
Determination of transfection efficiency Six hours after transfection with FAM-labeled siRNA-NC at various final concentrations, the cells were analyzed by FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA) equip-ped with Cell Quest Software (Becton Dickinson).
Real-time RT-PCR The total RNA was extracted from the LiBr cells using TRIzol reagent according to the manufacturer’s protocol. First-strand cDNA was synthesized using the ExScript RT-PCR reagent kit according to the manufacturer’s instructions.The specific primers for livin (forward: 5'-GTC AGT TCC TGC TCC GGT CAA-3', reverse: 5'-GGG CAC TTT CAG ACT GGA CCTC-3', 189 bp) and for GAPDH (forward: 5'-GCA CCG TCA AGG CTG AGAAC-3', reverse: 5'-ATG GTG GTG AAG ACG CCA GT-3', 142 bp) were designed and synthesized by TaKaRa. Real-time PCR was prepared using the SYBR premix Ex Taq kit according to the manufacturer’s protocol, and amplification was performed on an ABI Prism 7000 detection system (Applied Biosystems, Foster, CA, USA) according to the conditions recommended by the manufacturer, with an initial denaturation step at 95 ℃ for 10 s, followed by 40 cycles of denaturation at 95 ℃ for 5 s and 60 ℃ for 31 s. At the end of the amplification, a melting curve (disassociation curve) was run to ensure that only a single specific product was amplified. The data were analyzed by ABI Prism 7000 SDS software and the cycle threshold (Ct) values were determined. The modification of the 2–ΔΔCt method[12] was used to calculate changes of the relative expression of livin normalized against GAPDH.
Western blotting The total protein was extracted from the LiBr cells using RIPA lysis buffer and 1:100 dilution of a protease inhibitor cocktail (Sigma, St Louis, MO, USA). Western blotting was performed as described in a previous study[13] and modified. Thirty micrograms of the protein lysate was resolved electrophoretically on SDS-PAGE (12% for livin and 15% for caspase-3) and blotted onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). After being blocked for 1 h in blocking buffer (5% non-fat dried milk and 0.5% Tween-20 in TBS) and separately incubated with an antihuman livin mouse monoclonal antibody (1:1000), antihuman caspase-3 rabbit polyclonal antibody (1:500), and antihuman β-actin rabbit polyclonal antibody (1:3000) for 2 h at room temperature, the blots were washed 3 times with TBST (0.5% Tween in TBS) and incubated for 1.5 h with goat antimouse IgG/HRP (1:3000) or goat antirabbit IgG/HRP (1:3000) at room temperature, followed by washing 3 times with TBST. The signals were visualized with the enhanced chemiluminescence method and developed with X-ray film. The band density was measured by the GEL DOC 2000 system equipped with Quantity One software (Bio-Rad, Hercules, CA, USA) and normalized against the density of the respective housekeeping β-actin.
Terminal deoxynucleotidyl transferase-mediated digoxigenin-dUTP nick-end labeling assay The cells were seeded at 1.5×105 per well in 6-well plates with a coverslip for the seeded cells to grow on. At 72 h post-transfection, apo-ptotic cells were detected by terminal deoxynucleotidyl transferase-mediated digoxigenin-dUTP nick-end labeling (TUNEL) using the in situ cell death assay kit according to the instructions of a previous study[14], but with minor modifications: the cells were stained only with 3,3'-diamino-benzidine-tetrachloride and not counterstained. The apoptosis index was calculated as the percentage of cells with definite positive TUNEL staining and was obtained by counting 5 randomly chosen fields in each slide under a light microscope.
Flow cytometric analysis The LiBr cells were seeded at 7.5×104 per well in 12-well plates and transfected as described earlier. At 72 h post-transfection, the cells were harvested, stained with FITC-labeled Annexin V and propidium iodide (PI; Sigma, USA) to explore apoptosis on the FACSCalibur flow cytometer using Cell Quest software.
In addition, the cells were harvested, fixed in 70% ethanol for 12 h at 4 ℃, and stained with PI in a phosphate-buffered saline solution containing RNase (Roche, Basel, Switzerland) for the cell cycle analysis using Modfit software (Variety Software House, Topsham, ME, USA).
In vitro cell proliferation assay The cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. The LiBr cells were seeded at 5×103 per well in 96-well flat-bottom plates 1 d before transfection. After transfection for 24, 48, 72, and 96 h, followed by the addition of 20 µL of 5 mg/mL MTT (Sigma, USA) to each well, the cells were incubated for another 4 h, and 150 µL DMSO (Sigma, USA) was added and then lysed for 15 min. The absorbance value was measured on a microplate reader (Bio-Rad, USA) at 490 nm.
Statistical analysis The obtained data were statistically evaluated by ANOVA and presented as mean±SD from 3 independent experiments. Probability values of less than 0.05 were considered significant. All analyses were carried out using SPSS 10.0 statistical software (SPSS, Chicago, IL, USA).
Results
Expression of livin and transfection efficiency with siRNA in LiBr cells To identify whether LiBr cells express livin, Western blotting was performed. Unexceptionally, like other malignant melanoma cell lines, the LiBr cells expressed the livin protein obviously (data not shown).
To evaluate the transfection efficiency of LiBr cells with the siRNA duplexes, flow cytometric analysis was performed 6 h after transfection with FAM-labeled siRNA-NC and showed a significant transfection efficiency of 83.27%±4.11% at 75 nmol/L in a dose-dependent manner within 25–75 nmol/L (P<0.05). Thus, 75 nmol/L siRNA duplexes served as the initial concentration in the following experiments for the selection and validation of optimal siRNA sequences.
Knockdown of livin expression in LiBr cells by RNAi To select and validate effective siRNA target sites and determine the optimal dose- and time-response effect of silencing livin, the expression of livin in the LiBr cells that were separately transfected with siRNA-1, siRNA-2, or siRNA-3 at various concentrations was detected by real-time RT–PCR and Western blotting at 24, 48, 72, and 96 h post-transfection, respectively. Markedly, the silencing effects of siRNA-1, siRNA-2, or siRNA-3 on livin varied greatly (data not shown), and of them, only siRNA-3 achieved the highest silencing efficacy in dose- and time-dependent manners; 100 nmol/L of siRNA-3 with a maximum downregulation of 76.94%±6.33% on mRNA at 48 h and of 83.39%±5.44% on the protein at 72 h after transfection, respectively (partial data shown in Figure 1).
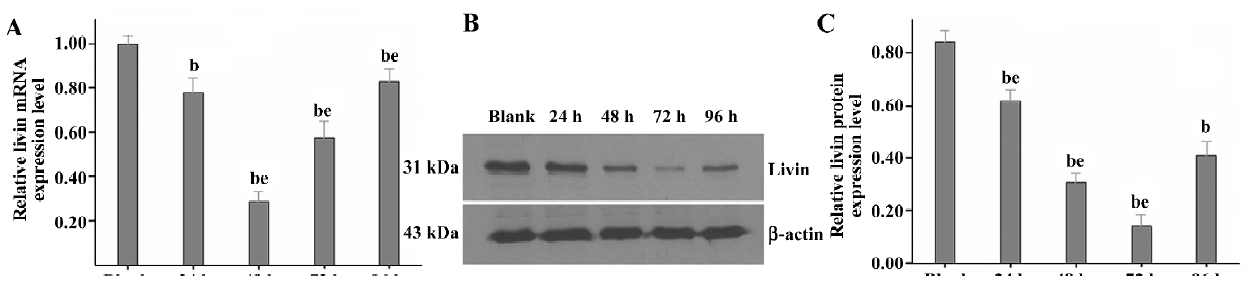
Therefore, siRNA-3 was selected as the effective siRNA sequence, and 100 nmol/L as the final concentration for use in investigating the effects of silencing livin on LiBr cells at 72 h post-transfection in the following experiments (except for the MTT assay).
Induction of apoptosis in LiBr cells by siRNA The TUNEL assay was performed to investigate whether silencing the livin gene induces apoptosis. The apoptosis index of cells transfected with siRNA-3 was 15.97%±2.56%, significantly higher than that of the cells in the blank (6.23%±1.12%), mock (6.47%±1.51%), and siRNA-NC (7.00%±1.11%; P<0.05) groups. In contrast, the apoptotic cells in the blank, mock, and siRNA-NC group did not differ from each other (P>0.05; Figure 2A).
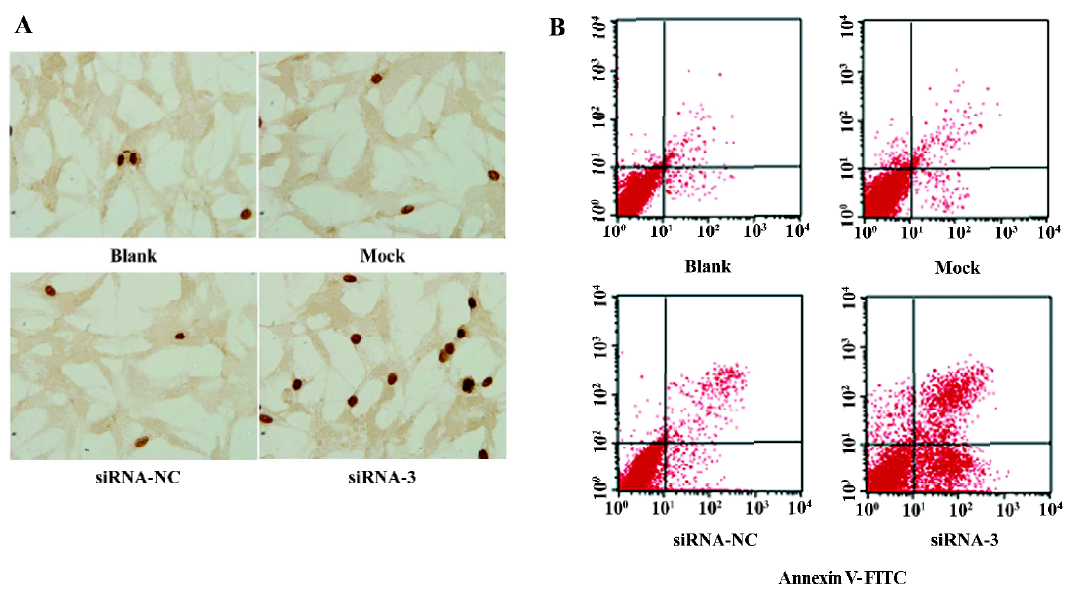
To further confirm the induction of apoptosis and quantify the apoptotic cells by silencing livin, flow cytometric analysis was performed. The early apoptotic rate of cells transfected with siRNA-3 increased to 28.67%±5.55% and the late apoptotic rate increased to 12.91%±3.77%, which were significantly greater than those in of the cells in the blank, mock, and siRNA-NC groups (P<0.05); there were no significant differences in the apoptotic cells among the blank, mock, and siRNA-NC groups (P>0.05; Figure 2B).
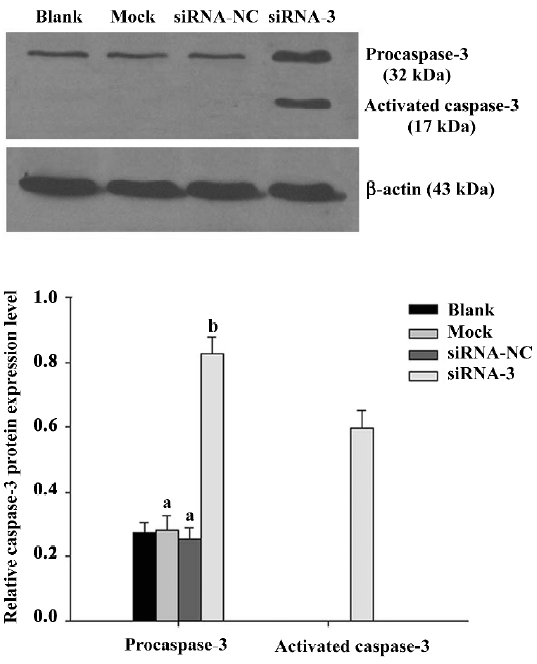
To explore whether silencing livin activates caspase-3 in LiBr cells, the expression of procaspase-3 and activated caspase-3 was analyzed by Western blotting. The protein level of procaspase-3 was obviously downregulated (P<0.05) and the cleaved form of caspase-3 was found in cells transfected with siRNA-3 compared with the blank control. In contrast, the levels of procaspase-3 were unchanged (P>0.05) and no cleaved fragment was detected in the cells of the blank, mock, and siRNA-NC groups (Figure 3).
Cell cycle arrest in LiBr cells by siRNA To examine whether silencing livin leads to cell cycle arrest, the phase distribution of the cell cycle was analysed by flow cytometry. Compared with the blank, mock, and siRNA-NC groups, there were great changes of cell cycle distribution in the cells transfected with siRNA-3; the cells blocked in the G0/G1 phase increased to 69.41%±4.41% (P<0.05), while the cells in the S phase decreased to 18.59%±2.65% (P<0.05). In contrast, there were no notable differences in the cell cycle distribution among the blank, mock, and siRNA-NC groups (P>0.05) (Figure 4A).
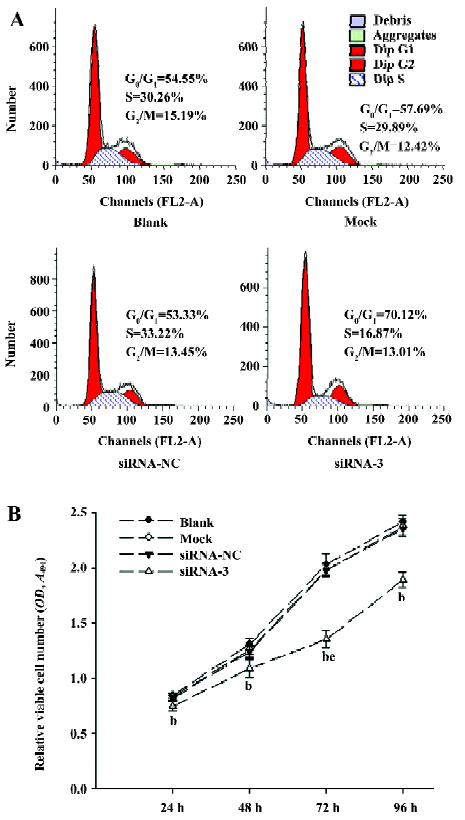
Inhibition of the proliferation in LiBr cells by siRNA The MTT assay showed that, compared with the blank group, the proliferation of cells transfected with siRNA-3 was remarkably inhibited from 24 to 96 h (P<0.05), with the highest inhibitory rate of 33.35%±3.46% (P<0.05 vs 24, 48, and 96 h groups) at 72 h post-transfection, but at 96 h, the inhibitory rate decreased to 21.82%±3.20%, which indicated that the number of viable cells began to increase. In contrast, there was no obvious difference in cell proliferation among the blank, mock, and siRNA-NC groups (P>0.05; Figure 4B).
Discussion
As resistance to apoptosis is a hallmark of various cancers and may be the underlying basis for tumorigenesis and tumor progression, strategies of inducing cancer cells to apoptosis are being designed[15,16]. Caspases are critical for the induction of apoptosis and their decreased expression is correlated with an increased grade of cancer, while the increased expression of caspases renders the cancer cells susceptible to chemotherapy[17,18]. However, the endogenous functions of caspases are inhibited by IAP that bind activated caspases in cancer cells[19]. Thus, removing the negative effects of the IAP represents a promising strategy to sensitize cancer cells to apoptosis[15]. RNAi technique can be applied to increase the apoptotic susceptibility of cancer cells by knocking down anti-apoptotic genes, such as certain IAP that were overexpressed. IAP seem to fit ideally as a specific molecular target because they are differentially overexpressed in many cases of malignant cells, but not in their healthy counterparts, and act at the effector level of the apoptosis pathways[15]. Up until now, 8 IAP have been identified in human cells, and among them, livin has been recently identified[4]. Previous reports have indicated that livin was overexpressed in a variety of malignancies and especially in malignant melanoma cells[1,2,5,6]. Considering that malignant melanoma, the most dangerous form of skin cancer, is resistant to currently available therapeutics[20], silencing the livin gene may be an encouraging approach for the gene therapy of malignant melanoma.
Our present study demonstrates that the expression of livin in LiBr cells could be knocked down specifically and effectively by siRNA in dose-and time-dependent manners. The silencing efficiency of the 3 designed specific siRNA duplexes that were separately targeted to the sites upstream (294–312), midstream (541–559), and downstream (790–808) of livin mRNA demonstrated striking differences. Only siRNA-3 resulted in a significant downregulation of livin expression. This phenomenon was believed to be associated with the positional effects[21].
In this study, we observed that silencing livin could notably promote apoptosis in LiBr cells. In accordance with the observations of increasing apoptotic cells, the cleaved form of caspase-3 (17 kDa) was found and the expression of procaspase-3 protein was upregulated. IAP are able to inhibit apoptosis by direct binding and the inhibition of certain caspases through 1 or more repeats of a highly-conserved 70 amino acid domain termed the baculovirus IAP repeat (BIR) domains. Livin with only 1 BIR domain is also able to inhibit caspases-3, -7, and -9[15]. Caspases form the core activation cascade of apoptosis with upstream or initiator caspases-8, -9, and -10, and downstream or effector caspases-3, -6, and -7. Caspase-3 is the effector protein of both the intrinsic and extrinsic pathways of apoptosis initiation[15]. The cleaved form of caspase-3 induced by siRNA indicated that silencing livin released caspase-3 from negative regulation by livin to activation and triggered apoptosis in the LiBr cells. This is in agreement with a previously published study on HeLa cells[22].
Our results demonstrated for the first time that silencing livin by siRNA leads to cell cycle arrest at the G0/G1 phase. In addition to the suppression of apoptosis, cancer cells are also characterized by deregulated cell proliferation, which is generally associated with accelerated G1/S and G2/M cell cycle transitions[23]. Some signals of DNA damage lead to cell cycle arrest at the G1 and/or G2 phases, reducing the rate of DNA synthesis or resulting in apoptosis[24,25]. The checkpoint responses and induction of apoptosis are considered to be major mechanisms for reducing both the initiation and progression of cancer[26,27]. In this study, the accumulation in the G0/G1 phase and the reduction in the S phase of LiBr cells treated with siRNA-3 indicated that silencing the livin gene resulted in a decrease of the ability of cells to progress from the G0/G1 to the S phase and synthesize DNA, eventually resulting in the inhibition of proliferation, which was confirmed by MTT assay. The study of cell proliferation with time-courses showed that the highest inhibitory rate was accordance with the maximum of the livin protein being downregulated at 72 h post-transfection. In this study, we provide preliminary evidence that silencing livin can change the cell cycle in LiBr cells. The results of a previous study indicated that livin might be regulated by cell cycle proteins similar to survivin based on the observation that livin and survivin have a similar subcellular localization[1]. Whether livin acts as a bifunctional protein associated with the regulation of the cell cycle and the inhibition of apoptosis-like survivin needs to be further explored.
In conclusion, silencing the livin gene by siRNA can significantly knock down the expression of livin, induce apoptosis, arrest the cell cycle at the G0/G1 phase, and inhibit proliferation in LiBr cells. Livin can serve as a potential molecular target to malignant melanoma in gene therapy by siRNA. The validated sequence of siRNA-3 can be used to construct vectors for stable expression in future.
Acknowledgements
We are grateful to Professor Xin-yang WANG for his helpful technical advice and skillful technical assistance. We thank Professor Tian-wen GAO for his kind gift of the malignant melanoma cell line, LiBr. We also thank Professor Dian-zen ZHANG, Professor Xiao-hui FAN, Dr Xian-fu YANG, and Dr Su CUI for their valuable help.
References
- Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem 2001;276:3238-46.
- Vucic D, Stennicke HR, Pisabarro MT, Salvesen GS, Dixit VM. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr Biol 2000;10:1359-66.
- Lin JH, Deng G, Huang Q, Morser J. KIAP, a novel member of the inhibitor of apoptosis protein family. Biochem Biophys Res Commun 2000;279:820-31.
- Liu B, Han M, Wen JK, Wang L. Livin/ML-IAP as a new target for cancer treatment. Cancer Lett 2007;250:168-76.
- Nachmias B, Ashhab Y, Bucholtz V, Drize O, Kadouri L, Lotem M, et al. Caspase-mediated cleavage converts Livin from an antiapoptotic to a proapoptotic factor: implications for drug-resistant melanoma. Cancer Res 2003;63:6340-9.
- Gong J, Chen N, Zhou Q, Yang B, Wang Y, Wang X. Melanoma inhibitor of apoptosis protein is expressed differentially in melanoma and melanocytic naevus, but similarly in primary and metastatic melanomas. J Clin Pathol 2005;58:1081-5.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494-8.
- Izquierdo M. Short interfering RNAs as a tool for cancer gene therapy. Cancer Gene Ther 2005;12:217-27.
- Li CY, Gao TW, Wang G, Han ZY, Li TH, Liu YF. The effect of antisense tyrosinase-related protein 1 on melanocytes and malignant melanoma cells. Br J Dermatol 2004;150:1081-90.
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol 2004;22:326-30.
- Ashhab Y, Alian A, Polliack A, Panet A, Ben Yehuda D. Two splicing variants of a new inhibitor of apoptosis gene with different biological properties and tissue distribution pattern. FEBS Lett 2001;495:56-60.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001;25:402-8.
- Du Y, Yin F, Liu C, Hu S, Wang J, Xie H, et al. Depression of MAD2 inhibits apoptosis of gastric cancer cells by upregulating Bcl-2 and interfering mitochondrion pathway. Biochem Biophys Res Commun 2006;345:1092-8.
- Zhang HZ, Wang Y, Gao P, Lin F, Liu L, Yu B, et al. Silencing stathmin gene expression by survivin promoter-driven siRNA vector to reverse malignant phenotype of tumor cells. Cancer Biol Ther 2006;5:1457-61.
- Nachmias B, Ashhab Y, Ben-Yehuda D. The inhibitor of apoptosis protein family (IAPs): an emerging therapeutic target in cancer. Semin Cancer Biol 2004;14:231-43.
- Fischer U, Schulze-Osthoff K. New approaches and therapeutics targeting apoptosis in disease Pharmacol Rev 2005;57:187-215.
- Kumar S. Caspase function in programmed cell death. Cell Death Differ 2007;14:32-43.
- Kumar MV, Shirley R, Ma Y, Lewis RW. Role of genomics-based strategies in overcoming chemotherapeutic resistance. Curr Pharm Biotechnol 2004;5:471-80.
- Shi Y. Caspase activation, inhibition, and reactivation: a mechanistic view. Protein Sci 2004;13:1979-87.
- Namkoong J, Martino JJ, Chen S. From existing therapies to novel targets: a current view on melanoma. Front Biosci 2006;11:2081-92.
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res 2002;30:1757-66.
- Crnkovic-Mertens I, Hoppe-Seyler F, Butz K. Induction of apoptosis in tumor cells by siRNA-mediated silencing of the livin/ML-IAP/KIAP gene. Oncogene 2003;22:8330-6.
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature 2001;411:342-8.
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004;73:39-85.
- Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology 2002;181–182:475-81.
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature 2000;408:433-9.
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication check-points. Annu Rev Genet 2002;36:617-56.
