Inhibition of β-lactamase-mediated oxacillin resistance in Staphylococcus aureus by a deoxyribozyme1
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) remains a major cause of nosocomial disease in the world, causing 50% or more of hospital-acquired S aureus infections in several countries[1]. In addition, recent reports have indicated that the epidemiology of MRSA may be undergoing a change through the emergence of community-acquired MRSA (CA-MRSA)[2,3]. CA-MRSA is capable of causing infections in otherwise healthy people and may have a serious or even fatal outcome[4,5]. According to the World Health Organization, more than 60% of S aureus strains worldwide are now resistant to methicillin. The mortality from severe MRSA infection is reported to be as high as 10%?34%[6,7].
Infections caused by MRSA are not effectively treated by most antibacterial agents and are a major challenge for chemotherapy because these bacteria show resistance to all β-lactam antibiotics. MRSA has traditionally been treated with glycopeptides, such as vancomycin. However, the emergence of vancomycin-intermediate or -resistant Staphylococci has spurred renewed efforts in the discovery of new targets in MRSA for novel antibacterial agents with mechanisms radically different from existing compounds[8].
S aureus resist the attack by β-lactam antibiotics in 2 ways: by producing β-lactamase, which inactivates the β-lactam antibiotics, and by expressing new transpeptidases, which are impervious to antibiotic activity. Two genes, blaZ encoding β-lactamase and mecA encoding the penicillin-binding protein PBP2a, render S aureus resistant to antibiotics, respectively[9]. The transcription of these genes is regulated by transmembrane signal sensor/transducer proteins (BlaR1 and MecR1) and their partner repressor proteins (BlaI and MecI). When a β-lactam antibiotic binds to the extracellular sensor domain of BlaR1, a conformational change within BlaR1 leads to autocatalytic activation of the integral-membrane metalloprotease domain. The active protease facing the cytosol specifically cleaves BlaI, directly or indirectly, which subsequently initiates blaZ expression. The genes for signal transducers and repressors are contained in a gene operon (blaR1-blaI) that is divergently transcribed from its regulated gene blaZ[10]. This system highlights the key role of the methicillin sensor-transducer as the eventual transcriptional regulator of MRSA response[11,12]. Therefore, blocking the sensor/transducer pathway may represent a novel approach to reverse antibiotic resistance in MRSA.
In recent years, catalytic nucleic acids were composed entirely of DNA, which have been generated by in vitro selection strategies. These molecules ideally combine the catalytic activity of ribozymes with the stability of oligo-deoxynucleotides. The typical deoxyribozyme or DNAzyme, known as the“10–23” model, is capable of cleaving a specific phosphodiester linkage between an unpaired purine (A, G) and a paired pyrimidine (C, U) under simulated physiological conditions. DNAzymes have considerable advantages over ribozymes in that they are easier to synthesize and less sensitive to chemical and enzymatic degradation than RNA-based reagents. They also exhibit greater catalytic efficiency than conventional hairpin and hammerhead ribozymes[13,14], so it is considered as an easily “drugable” tool to knock down target genes. In this study, we explored the use of a synthetic deoxyribozyme targeting blaR1 transcripts as a potential tool in inhibiting the expression of blaR1, blocking the signal pathway of blaR1-blaI-blaZ and thereafter leading to the reduction of BlaZ expression. The PS-DRz602 (anti-blaR1 phosphorothioate deoxyribozyme) was found to reduce β-lactamase mRNA expression concomitantly and led to the restoration of the susceptibility of MRSA to oxacillin.
Materials and methods
Bacterial strain and electrocompetent S aureus preparation The strain of MRSA, WHO-2, obtained from Chinese National Center for Surveillance of Antimicrobial Resistance (Beijing, China) was used in the study. WHO-2 exhibited a moderate level of resistance to oxacillin (minimum inhibitory concentration [MIC]=32 μg/mL), in which the mecR1-mecA[15] and blaR1-blaZ[16] genes were detected by PCR. A methicillin-susceptible S aureus strain, ATCC (American Type Culture Collection) 29213, was used as a positive control.
Competent S aureus was prepared according to a previously published paper[17]. Briefly, 2 mL of overnight-cultured WHO-2 was transferred to 200 mL broth medium and incubated at 37 oC with moderate agitation until the optical density (OD)600 reached 0.55-0.65. The cells were centrifuged at 2817×g for 10 min at 4 oC and the supernatant was removed. The pellet was washed by resuspending in ice-cold deionized water in the same volume of culture medium and the suspension was centrifuged. The supernatant was carefully removed and the pellet was washed a second time using the same procedure. The pellet was then washed 4 times with 40, 10, 2, and 1 mL of 10% cold glycerol, respec-tively, with the procedure mentioned above. Finally, the cell pellet was resuspended in 1 mL of cold 10% glycerol and 50 μL aliquots and stored at -80 oC.
Modified DNAzyme The sequence of most active DNAzyme in this study was PS-DRz602: 5'-
DNAzyme delivery Different concentrations of phos-phorothioated PS-DRz602 (5, 10, and 15 mg/L) were introduced into the S aureus strain WHO-2 by the Electroporator (JY2000-1B electroporation apparatus, Ningbo Scientz Biotechnology, Ningbo, China) at conditions of 25 µf, 900 V, 200 Ω, and time constant 3.6-4.2 ms. Briefly, the cells of bacteria and PS-DRz602 were mixed and transferred into prechilled cuvettes. After the pulses were applied, the cuvettes were removed and 1 mL of SOC (Super Optimal Catabolite) medium (0.5% yeast extract, 2% tryptone, 10 mmol/L NaCl, 2.5 mmol/L KCl, 10 mmol/L MgCl2, 20 mmol/L MgSO4, and 20 mmol/L glucose [pH 7.0]) was added immediately. The cells were allowed to recover by incubating for 1 h at 37 oC with shaking. Transformation efficiencies should be approximately 9×108 transformants/µg of DNAzyme.
Bacterial susceptibility assay The growth determination of the cells receiving different concentrations of PS-DRz602 by electroporation was carried out as follows: the cells were diluted 106 times and 50 µL dilution was then spread onto the Mueller-Hinton agar that contained 6 mg/L oxacillin; the plates were incubated for 48 h at 35 oC. The number of colonies were counted for plates with >10 and <500 colonies, and the total colony-forming unit (CFU) per sample was determined by correcting the colony count for the dilution.
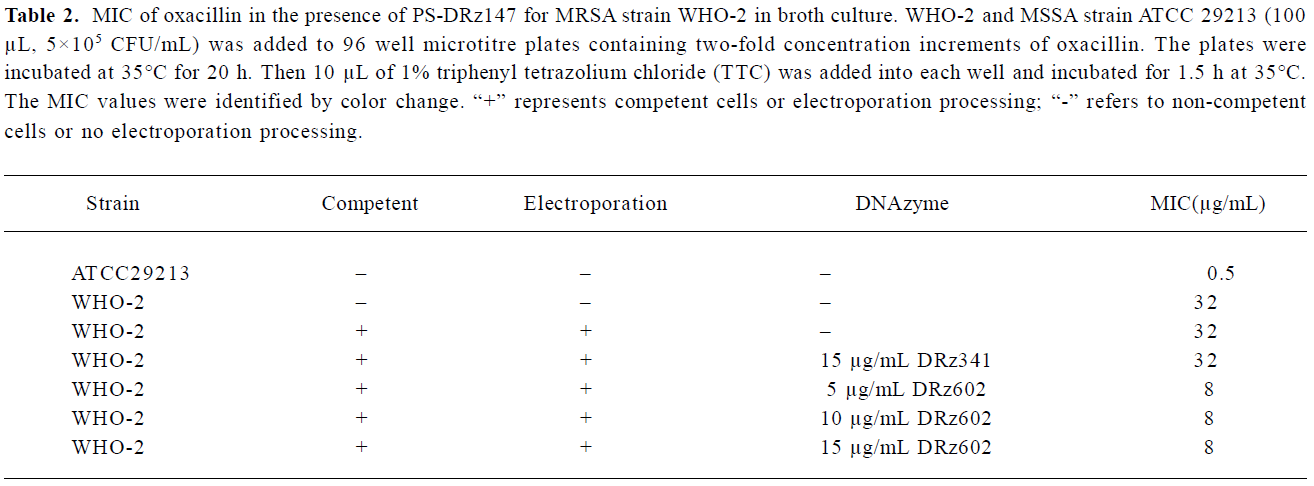
The MIC of oxacillin for PS-DRz602-treated MRSA and MSSA (methicillin sensitive S aureus) were determined by the 2-fold microdilution method with the Mueller-Hinton broth supplemented with oxacillin in the range of 0.25-128 µg/mL. 100 µL of 5×105 CFU/mL test bacteria was added to 96-well microtitre plates and grown at 35 °C for 20 h. 10 µL of 1% triphenyl tetrazolium chloride (TTC), a colorimetric indicator, was added to each well of the microtitre plates and incubated for 1.5 h at 35 °C. The TTC-based MIC was determined as being the lowest concentration of oxacillin that showed no red color changes and indicated the complete growth inhibition.
RNA extraction The culture of the S aureus strain WHO-2 was centrifuged at 1957×g for 10 min at 4 oC and the supernatant was decanted. The cell pellet was suspended in 100 µL lysis solution (50 mmol/L Tris-HCl [pH 7.5], 5 mmol/L EDTA [pH 8], and 50 mmol/L NaCl) with 300 µg lysozyme (Sigma-Aldrich, St Louis, MO, USA) and 5 µg lysostaphin (Sigma-Aldrich, USA) for 30 min at room temperature. The total RNA was extracted from the bacterial lysis with Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacture’s instructions, and the RNA samples were treated with DNase I to remove any genomic DNA contamina-tion.
Reverse transcription reaction The cDNA of blaR1 and blaZ was synthesized, respectively, by reverse transcription (RT) from 1 µg of each RNA sample using SuperScriptIII reverse transcriptase (Invitrogen, USA). The 14 µL mixture (1 µg RNA, 0.1 µg random primer, 4 µL of 2.5 mmol/L dNTP mix, and sterile water) was heated at 65 °C for 5 min and incubated on ice for at least 1 min. 1 µL of 0.1 mol/L DTT (dithio-threitol), 4 µL of 5× RT buffer, 20 U RNase inhibitor, and 100 U SuperScriptIII reverse transcriptase was added to the mixture in a final volume of 20 µL. The RT conditions were 5 min at 25 °C, 45 min at 50 °C, and 15 min at 70 °C.
Real-time PCR detection The nucleotide sequences for the various primers are listed in Table 1. All the primers were synthesized commercially (Shanghai Sangon Biological Engineering Technology and Services, China).
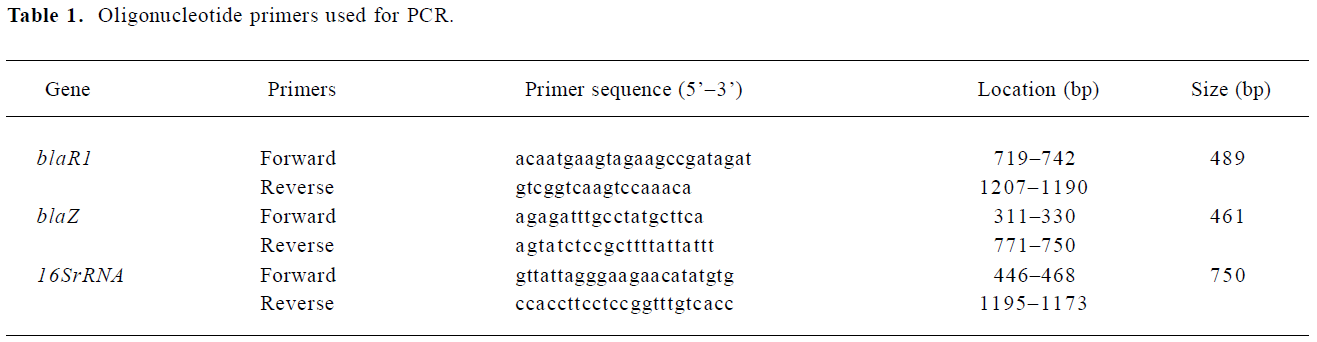
Full table
The PCR was run in a DNA Engine Opticon (MJ Research, Waltham, MA, USA) with SYBR Green I. The PCR reagents consisted of: 12.5 µL SYBR Premix Ex Taq (DRR041S, TaKaRa, Otsu, Shiga, Japan), 0.5 µL of 50×OX reference dye (DRR041S, TaKaRa, Japan), 0.75 µL of each primer (10 µmol/L), and 1 µL of sample cDNA in a final volume of 25 µL. Each plate included its own negative controls: no template controls (where all the reaction reagents except for cDNA were used). The thermal cycling conditions were: an initial denaturation step at 95 °C for 5 min, then 50 cycles at 95 °C for 10 s, 55 °C for 20 s, and 72 °C for 20 s. The melting curves of the PCR products were acquired by the stepwise increase of the temperature from 60 to 90 °C (temperature transition 0.5 °C/s). The standard deviation of the fluorescence values recorded from cycles 3 to 15 was multiplied by 10 to define the cycle threshold line. The specificity of the amplified products was verified by analysis of the dissociation curves as well as by ethidium bromide-stained 1% agarose gels.
The DNA Engine Opticon system detects and plots the increase of each PCR product in fluorescence versus the PCR cycle number to produce a continuous measurement of PCR amplification. To provide the precise quantification of the initial target in each PCR reaction, the amplification plot was examined at a point during the early log phase of product accumulation. This was accomplished by assigning a fluorescence threshold above the background and determining the time-point at which each sample’s amplification plot reached the threshold (defined as the threshold cycle number or Ct). Differences in the threshold cycle number were used to quantify the relative amount of PCR target contained within each tube.
Construction of standard curves for blaR1, blaZ, and 16SrRNA genes The cDNA of control group was 10 fold series diluted and the analysis was performed as follows: For each sample, a difference in Ct values (ΔCt) was calculated for target genes (blaR1/blaZ) by taking the mean Ct of duplicate tubes and subtracting the mean Ct of the duplicate tubes for the reference RNA (16SrRNA) measured on an aliquot from the same RT reaction. ΔCt =Ct (target gene)-Ct (16SrRNA).
Comparative calculation and determination of the relative expression levels of blaR1 and blaZ in the differently treated groups The relative expression of blaR1 or blaZ mRNA was calculated using the comparative Ct method. Real-time relative quantitations of blaR1 and blaZ expressions were performed as previously described[18]. The 16SrRNA gene, which was expressed at relatively the same level throughout the developmental cycle in WHO-2, was used as the control to normalize the quantity of a cDNA target to determine differences in the amount of total cDNA in a reaction[18,19]. The ΔΔCt values were calculated as the following equation: ΔΔCt =ΔCt (treatment)-Ct (control). The ΔCt for the treated sample was then subtracted from the ΔCt for the untreated control sample to generate ΔΔCt .
The mean of these ΔΔCt measurements was then used to calculate the expression of the test gene (2-ΔΔCt) relative to the reference gene and normalized to the untreated control as follows: Relative expression=2-ΔΔCt. The evaluation of 2-ΔΔCt indicates the fold change in gene expression relative to the untreated control.
Statistical analysis Values are expressed as mean±SD and one-way ANOVA analysis followed by SNK (Student-Newman-Keuls) t-test was performed. P<0.05 was considered statistically significant.
Results
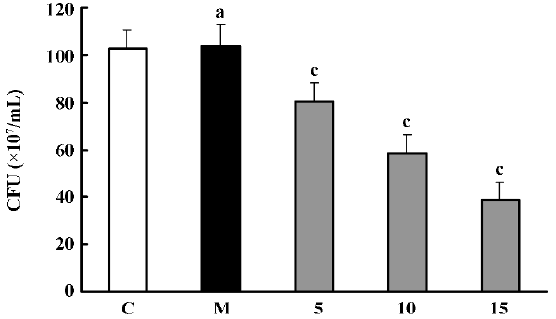
Effects of PS-DRz602 on colony forming of the WHO-2 strain The number of WHO-2 colonies on the Mueller-Hinton agar containing oxacillin (6 mg/L) was significantly decreased to 78.2%, 56.7%, and 37.8% of the control value in all anti-BlaR1 PS-DRz602-treated groups concentration-dependently. However, the growth of WHO-2 was neither influenced in the mismatched PS-DRz341-treated group nor affected in the control group without PS-DRz602 treatment (Figure 1).
Partial restoration of antibiotic susceptibility in the MRSA strain WHO-2 We found that the down-regulation of blaR1 by the introduction of anti-blaR1 PS-DRz602 can partially increase the susceptibility of WHO-2 to oxacillin (Table 2). The MIC of oxacillin was reduced from 32 to 8 µg/mL in the presence of 5, 10, and 15 µg/mL anti-blaR1 PS-DRz. In contrast, the MIC of oxacillin for mismatched PS-DRz341 not targeting blaR1 remained unchang-ed. Competence or electroporation alone did not affect the MIC of oxacillin on WHO-2 (Table 2).
Full table
Real-time quantitation assays for blaR1 and blaZ transcription We next determined whether the conversion of antibiotic resistance to antibiotic susceptibility in MRSA strain WHO-2 was accompanied by the inhibition of blaR1 and blaZ mRNA expression through anti-blaR1 PS-DRz602.
An analysis of the melting curves of the PCR products for blaR1, blaZ, or 16SrRNA showed a single-peak graph for all amplifications, indicating that a single PCR product was formed. This was confirmed by running 10 µL of each product on an ethidium bromide-stained 1% agarose gel.
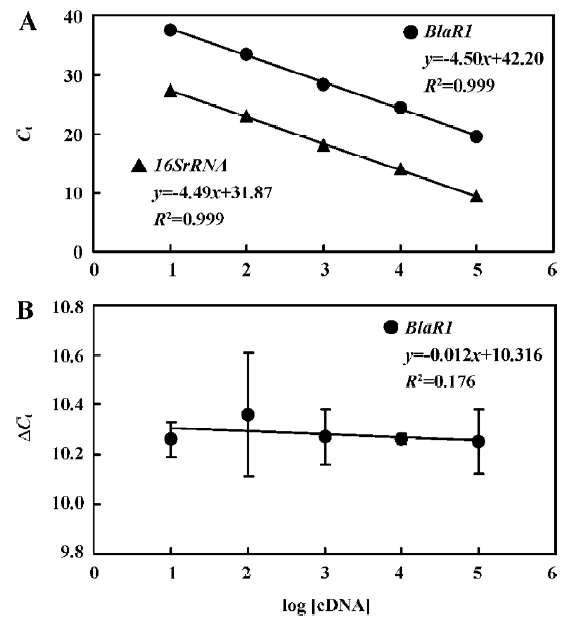
The standard curves for blaR1 or blaZ and 16SrRNA were generated using cDNA from WHO-2 (Figures 2, 3). To demonstrate that the PCR efficiencies for the target and the control genes were approximately equal, the values of the 2 standard curves were used to determine the absolute value of the slope of the log of cDNA for each dilution versus ΔCt (difference in the cycle threshold obtained for the 2 PCR systems for the same cDNA dilution) for the respective dilution. This validation experiment involved pairwise comparisons between blaR1 and 16SrRNA or blaZ and 16SsRNA. The slope was 0.012 and 0.010 for each experiment, respec-tively, indicating approximately equal amplification efficiencies between blaR1 or blaZ and 16SrRNA (Figures 2, 3).
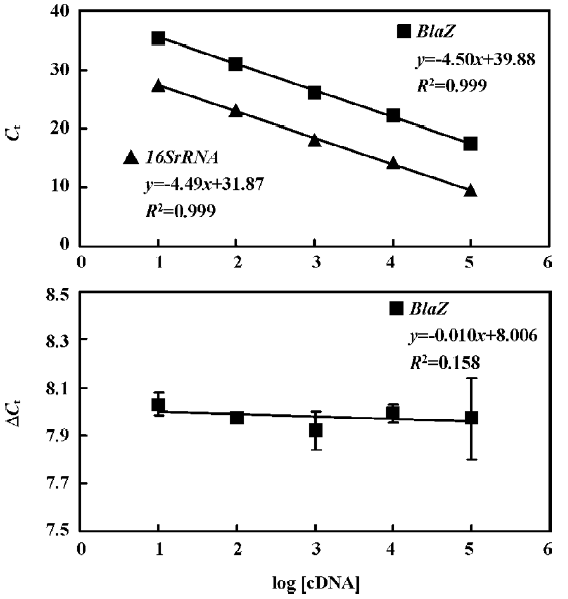
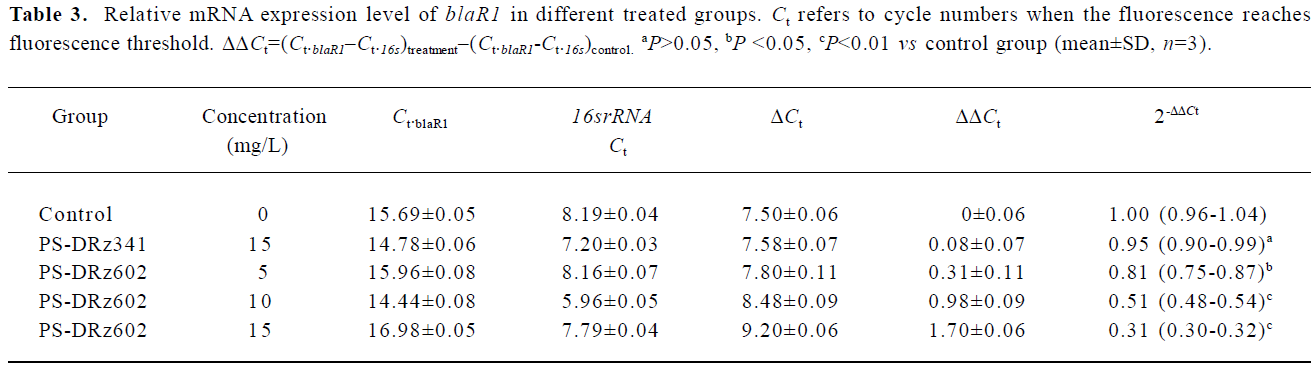
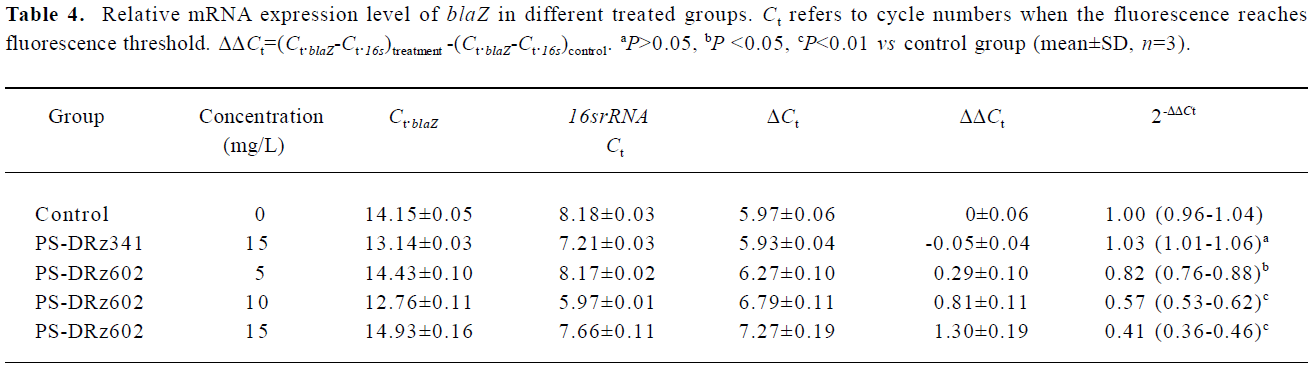
Comparative transcription of a single gene for blaR1 and blaZ The change in the transcription of the target genes (blaR1 and blaZ) normalized to 16SrRNA was monitored in the different concentration groups. As expected, the transcriptional change of blaR1 in the differently treated groups was coincidental to that of blaZ. Compared with the control group, the relative transcription of blaR1 in 3 groups (5, 10, 15 mg/L PS-DRz602) was decreased to 81%, 51%, and 31% of the control values, respectively, in a concentration-dependent manner (Table 3). Interestingly, the blockade of the transcription of blaR1, a sensor transducer gene, led to the concentration-dependent repression of its downstream gene blaZ to 82%, 57%, and 41% of the control values, respectively(Table 4).
Full table
Full table
Discussion
Because of the emergence and spread of resistance genes[9-12,20], MRSA has been the cause of major outbreaks and epidemics among hospitalized patients, with high mortality and morbidity rates. Recently, some schemes have shown both a dramatic rise in the total numbers of cases of S aureus bacteremia reported annually and an increase in the proportion of such cases that involve MRSA (from 2% in 1990 to >40% in the early 2000s)[21]. However, the emergence of vancomycin-resistant MRSA and treatment failure of MRSA infections has led to the urgent need for alternative anti-MRSA therapies.
The different antisense approaches have demonstrated the feasibility of using antisense oligonucleotides or oligonucleotide analogs in the treatment of bacterial infections, and the promising results have been observed by researchers in vitro and in vivo[22,23]. Sarno et al demonstrated that selectively-designed antisense oligonucleotides could bind to AAC(6')-I-type acetyltransferase (aminoglycoside 6'-N-acetyltransferase type Ib) mRNA, mediate RNase H digestion, and thereafter decrease the level of resistance to amikacin in Escherichia coli[24]. The application of peptide-PNA (peptide nucleic acid) conjugates, antisense phosphorodiamidate morpholino oligomer, or antisense DNA analogs showed a significant inhibition of gene expression in E Coli, respec-tively, and effectively reversed the multiple drug resistance of E coli[25-27]. When PS-ODN (phosphorothioate oligo-deoxynucleotide), targeting a specific region of the mycobacterium aspartokinase (ask) gene in Mycobacterium smegmatis were utilized in combination with ethambutol, the results showed a significant inhibition of growth of drug-resistant strains of M smegmatis[28]. The inhibition of glutamine synthetase activity with antisense oligonucleotides to glutamine synthetase mRNA also held back the replication of Mycobacterium tuberculosis[29]. Resistance to β-lactam was reversed by the blockade of the expression of bla in the presence of antisense peptide nucleic acid in E coli[30] and resistance to vancomycin was fully reversed when an Enterococcus faecalis isolator harbored recombinant shuttle vectors containing a vanH promoter-vanA antisense gene cassette[31]. The peptide nucleic acid treatment was effective in rescuing 100% of infected animals[23]. Antisense antibiotics have been proposed as a new hope for bacteria infection therapy through targeting specific genes in bacteria[32].
Our previous study indicated that a phosphorothioate oligodeoxynucleotide ODN6087 could inhibit mecR1 and mecA expression and partially restore the susceptibility of a MRSA strain[33]. The current study demonstrated that PS-DRz602 targeted to the sequence of blaR1 mRNA significantly inhibited the resistance of MRSA strain WHO-2 to oxacillin, together with the reduction of blaZ mRNA expression. The colony forming of WHO-2 was decreased to 78.2% (5 mg/L), 56.7% (10 mg/L), and 37.8% (15 mg/L) of the control values, respectively, in a concentration-dependent manner after PS-DRz602 treatment. However, anti-blaR1 PS-DRz602 had limited activity to reverse the susceptibility of MRSA strain WHO-2 in the present study. The reason for the insufficient restoration of susceptibility is that we delivered the PS-DRz to competent S aureus only once by electro-poration. Although the delivery efficiency was estimated at around 9?108 transformants/micrograms in all groups, PS-DRz was not retained in the culture medium constantly and bacterial proliferation diluted the PS-DRz. As demonstrated, the MIC of oxacillin to WHO-2 were reduced from 32 to 8µg/mL, which was still 2-folds higher than the margin value (2 µg/mL) of oxacillin sensitivity to S aureus and the inhibitory efficiency of the anti-blaR1 PS-DRz602 at 78.2%?37.8% of control values. However, if the highly-efficient delivery system is applied and the antisense PS-DRz is sustainable throughout proliferation, then a high efficiency of anti-blaR1 PS-DRz will be achieved.
Since BlaR1 is an upstream regulatory element for the initiation of BlaZ expression via the inactivation of BlaI, it was found that BlaZ expression was consequently greatly inhibited after the blockade of BlaR1. Although there is no direct evidence to prove the cleaving activity of PS-DRz602 to the mRNA substrates of BlaR1 precisely in WHO-2, the mismatched anti-blaR1 PS-DRz341 had no effects on the growing of WHO-2, as well as no influence on the expression of BlaR1, and β-lactam mRNA would provide indirect evidence to support the selectivity of PS-DRz602. In addition, in the cytoplasm of cells, a full length of mRNA often has a secondary structure. If the target sites of DNAzymes are within the secondary structure of the RNA, DNAzymes may still be ineffective[34,35]. Some studies have shown that DNAzymes work well in the in vitro system, but do not work in the whole cell[35,36]. Mitchell et al and Patzel et al found that without any prior screening, the initially synthesized DNAzyme sequence specifically reduced the target gene expression[34,37].
In conclusion, the results of our present study suggested that the blockade of the blaR1-blaZ signaling pathway via a DNAzyme was an alternative strategy to the reverse phenotype of antibiotic resistance of MRSA, and BlaR1 may be an attractive target for antimicrobial agent development.
Acknowledgement
We are grateful to Prof Yue MA (Chinese National Center for Surveillance of Antimicrobial Resistance, Beijing, China) for providing the WHO-2 strain.
References
- Aires de Sousa M, de Lencastre H. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol Med Microbiol 2004;40:101-11.
- Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis 2005;5:275-86.
- Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying panton-valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 2003;9:978-84.
- Eady EA, Cove JH. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus — an emerging problem for the management of skin and soft tissue infections. Curr Opin Infec Dis 2003;16:103-24.
- Crum NF. The emergence of severe, community-acquired methicillin-resistant Staphylococcus aureus infections. Scand J Infec Dis 2005;37:651-6.
- Tumbarello M, de Gaetano Donati K, Tacconelli E, Citton R, Spanu T, Leone F, et al. Risk factors and predictors of mortality of methicillin-resistant Staphylococcus aureus (MRSA) bacterae-mia in HIV-infected patients. J Antimicrob Chemother 2002;50:375-82.
- Crowcroft NS, Catchpole M. Mortality from methicillin resistant Staphylococcus aureus in England and Wales: analysis of death certificates. BMJ 2002;325:1390-1.
- Srinivasan A, Dick JD, Perl TM. Vancomycin resistance in Staphylococci. Clin Microbiol Rev 2002;15:430-8.
- Archer GL, Bosilevac JM. Signaling antibiotic resistance in . Science 2001; 291: 1915?6.
- Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 2003;111:1265-73.
- Safo MK, Zhao Q, Ko TP, Musayev FN, Robinson H, Scarsdale N, . Crystal structures of the BlaI repressor from and its complex with DNA: insights into transcriptional regulation of the bla and mec operons. J Bacteriol 2005; 187: 1833?44.
- Golemi-Kotra D, Cha JY, Meroueh SO, Vakulenko SB, Mobashery S. Resistance to beta-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in . J Biol Chem 2003; 278: 18 419?25.
- Emilsson GM, Breaker RR. Deoxyribozymes: new activities and new applications. Cell Mol Life Sci 2002;59:596-607.
- Khachigian LM. Catalytic DNAs as potential therapeutic agents and sequence-specific molecular tools to dissect biological function. J Clin Invest 2000;106:1189-95.
- Bignardi GE, Woodford N, Chapman A, Johnson AP, Speller DC. Detection of the mec-A gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J Antimicrob Chemother 1996;37:53-63.
- Yazdankhah SP, Sorum H, Oppegaard H. Comparison of genes involved in penicillin resistance in Staphylococci of bovine origin. Microb Drug Resist 2000;6:29-36.
- Augustin J, Gotz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett 1990;54:203-7.
- Eleaume H, Jabbouri S. Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J Microbiol Methods 2004;59:363-70.
- Goerke C, Bayer MG, Wolz C. Quantification of bacterial transcripts during infection using competitive reverse transcription-PCR (RT-PCR) and LightCycler RT-PCR. Clin Diagn Lab Immunol 2001;8:279-82.
- Dzidic S, Bedekovic V. Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta Pharmacol Sin 2003;24:519-26.
- Johnson AP, Pearson A, Duckworth G. Surveillance and epidemiology of MRSA bacteraemia in the UK. J Antimicrob Chemo-ther 2005;56:455-62.
- Chopra I. Prospects for antisense agents in the therapy of bacterial infections. Expert Opin Investig Drugs 1999;8:1203-8.
- Tan XX, Actor JK, Chen Y. Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob Agents Chemother 2005;49:3203-7.
- Sarno R, Ha H, Weinsetel N, Tolmasky ME. Inhibition of aminoglycoside 6'-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob Agents Chemother 2003;47:3296-304.
- Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol 2001;19:360-4.
- Geller BL, Deere J, Tilley L, Iversen PL. Antisense phosphoro-diamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J Antimicrob Chemother 2005;55:983-8.
- White DG, Maneewannakul K, von Hofe E, Zillman M, Eisenberg W, Field AK, et al. Inhibition of the multiple antibioc resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob Agents Chemother 1997;41:2699-704.
- Rapaport E, Levina A, Metelev V, Zamecnik PC. Antimyco-bacterial activities of antisense oligodeoxynucleotide phosphoro-thioates in drug-resistant strains. Proc Natl Acad Sci USA 1996;93:709-13.
- Harth G, Zamecnik PC, Tang JY, Tabatadze D, Horwitz MA. Treatment of Mycobaterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-L-glutamate/glutamine cell wall struture, and bacterial replication. Proc Natl Acad Sci USA 2000;97:418-23.
- Good L, Nielsen PE. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat Biotechnol 1998;16:355-8.
- Torres Viera C, Tsiodras S, Gold HS, Coakley EP, Wennersten C, Eliopoulos GM, et al. Restoration of vancomycin susceptibility in Enterococcus faecalis by antiresistance determinant gene transfer. Antimicrob Agents Chemother 2001;45:973-5.
- Geller BL. Antibacterial antisense. Curr Opin Mol Ther 2005;7:109-13.
- Meng JR, Hu BQ, Liu J, Hou Z, Meng J, Jia M, et al. Restoration of oxacillin susceptibility in methicillin-resistant Staphylococcus aureus by blocking the MecR1-mediated signaling pathway. J Chemother 2006;18:360-5.
- Patzel V, Steidl U, Kronenwett R, Haas R, Sczakiel G. A theoretical approach to select effective antisense oligodeoxyribonucleo-tides at high statistical probability. Nucleic Acids Res 1999;27:4328-34.
- Cairns MJ, Hopkins TM, Witherington C, Wang L, Sun LQ. Target site selection for an RNA-cleaving catalytic DNA. Nat Biotechnol 1999;17:480-6.
- Cairns MJ, Sun LQ. Target-site selection for the 10?23 DNAzyme. Methods Mol Biol 2004;252:267-77.
- Mitchell A, Dass CR, Sun LQ, Khachigian LM. Inhibition of human breast carcinoma proliferation, migration, chemoinvasion and solid tumour growth by DNAzymes targeting the zinc finger transcription factor EGR-1. Nucleic Acids Res 2004;32:3065-9.
