Inhibition of STAT3 expression by siRNA suppresses growth and induces apoptosis in laryngeal cancer cells
Introduction
Laryngeal carcinoma especially at late-stage is associated with high morbidity and poor long-term survival due to the absence of effective treatment methods. Better understanding of molecular mechanisms underlying proliferation, differentiation, and survival of laryngeal carcinoma is critical for the development of optimal therapeutic modalities. Recent studies suggest that signal transducers and activator of transcription (STATs) have the potential as novel molecular targets for the development and survival of laryngeal carcinomas.
STATs are latent cytoplasmic transcription factors that function as intracellular effectors of cytokine and growth factor signaling pathways . Among the STAT family, STAT3 plays a key role in promoting proliferation, differentiation, anti-apoptosis, or cell cycle progression. Constitutive activation of STAT3 is implicated in a variety of tumor cell lines[1–5], thereby suggesting that STAT3 is an important molecular target for tumor therapy.
In vitro studies have shown that inhibition of STAT3 activity in human tumor cells induces apoptosis and/or growth arrest. In human head and neck squamous carcinoma cells, blocking of STAT3 signaling by decoy oligonucleotide or antisense oligonucleotides abrogates transforming growth factor and suppresses oncogenic growth of these cells[6,7]. STAT3β is a naturally accruing dominant-negative STAT3 variant that is identical to STAT3 except for the absence of the transactivation domain[8]. Bowman et al[2] have reported that blockade of STAT3 signaling by STAT3β in human myeloma cells down-regulates IL-6-induced expression of the antiapoptotic gene, Bcl-xL, resulting in a dramatic sensitization of cells to Fas-mediated apoptosis in vitro. STAT3β can promote apoptosis in breast cancer cells, inhibit Bcl-xL expression, and induce apoptosis[9]. Overall, these raise the possibility that targeting STAT3 may enhance antitumor responses in vivo in a variety of human cancers.
A relatively new technique using RNA interference (RNAi) provides a novel approach of experimental inhibition of gene expression. RNAi is triggered by the presence of double-stranded RNA (dsRNA) in the cell and results in rapid degradation of the targeted mRNA with homology to the double strand leading to potent and selective silencing of genes. This phenomenon was first observed from studies in Caenorhabditis elegans and Drosophila melanogaster and subsequently, in other organisms[10]. Recent studies have shown that short interfering (21–25 bp) RNA molecules (siRNA), but not long dsRNA (greater than 30 bp), are key elements of RNAi. Only recently has the use of RNAi in mammalian studies been established by introducing siRNA[14]. At present, siRNA has been adapted as a functional genomic tool and has potential as a therapeutic approach in cancer.
Intriguingly, blocking of STAT3 signaling pathways by siRNA has been shown to suppress growth and induce apoptosis in prostate cancer cell lines and astrocytoma cells[15,16]. However, to date no studies have studied the effect of inhibition of STAT3 gene expression by siRNA on laryngeal cancer. Since STAT3 signaling is critical for the regulation of proliferation, differentiation, and apoptosis of tumor cells, we hypothesize that knockdown of STAT3 gene expression by siRNA should suppress tumor growths and induce apoptosis in laryngeal carcinoma that observed in models of prostate cancer and astrocytoma[16,17].
The objectives of the present study were (1) to determine the inhibitory effect of the synthetic STAT3 siRNA on the expression of STAT3 gene in laryngeal carcinoma cells and (2) to investigate the effect of STAT3 siRNA on the growths and apoptosis in laryngeal carcinoma cells.
Materials and methods
Construction of plasmids that contain DNA templates for the synthesis of siRNAs were constructed under the control of the U6 promoter The pSilencer1.0-U6 (Ambion Inc Austin, TX, USA) was used for DNA vector-based siRNA synthesis under the control of U6 promoter in vivo. In brief, first, the double stranded DNA template encoding siRNA oligonucleotides (GeneBank: access numbers for the human STAT3: NM003150) that contained a sense strand of 19 nucleotide sequences followed by a short space (TTCAA-GAGA), the reverse complement of the sense strand, and five thymidines as a RNA polymerase III transcriptional stop signal were synthesized. The sequences were forward 5´-GCAGCAGCTGAACAACATGTTCAAGAGACATGTTGT-TCAGCTGCTGCTTTTTT3´ and reverse 5´ AATTAAAAA-AGCAGCAGCTGAACAACATGTCTCTTGAACATGTTG-TTCAGCTGCTGCGGCC3´ (locate on SH2 domain). The oligo nucleotides were annealed in a buffer (potassium actate 100 mmol/L, 30 mmol/L HEPES-KOH pH 7.4, and magnesium acetate 2 mmol/L) and the mixture was incubated at 90 ºC for 3 min and then at 37 ºC for 1 h. The double stranded oligos were cloned into the ApaI–EcoR I sites of the pSilencer 1.0-U6 vector (Ambion Inc) where short hairpin RNAs (shRNA) were expressed under the control of the U6 promoter.
Cell culture and transfections The human laryngeal cancer cell lines Hep2 were obtained from ATCC. Hep2 cells were cultured in medium RPMI-1640 (Invitrogen, Inc Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and penicillin (100 kU/L) and streptomycin (100 mg/L) at 37 ºC in a humidified incubator with 5% CO2. For cell transfection, lipofectamine 2000 (Invitrogen) was used for transfecting the plasmids following the manufacturer instructions. In brief, pEGFP was cotransfected with pSilencer1.0-U6-siRNA-STAT3 or pSilencer empty vector at ratio of 1:20 to mark the positive transfected cells, respectively. The cells were cultured for 5–20 h and then transferred to fresh medium with 10% FBS and lysed for 24–72 h after transfection.
Northern blot Total RNA was extracted from cell samples with Trizol (Invitrogen, Carlsbad, CA, USA). Equal amounts of 20 µg total RNA were electrophoresed on 1.2% agarose gel with formaldehyde 2.2 mol/L and transferred onto nylon membranes (Hybond-N, Amersham Pharmacia Biotech). Blots were incubated with 32P-labeled cDNA against STAT3 and actin with Hyb and washed according to the manufacture directions. Visualization of blots was performed by overnight exposure to Kodak MS film. Quantification of blots developed on films was accomplished with a Molecular Dymanics phosphorimager.
Western blot Total protein was extracted from the harvested sample cells with protein lysis buffer (5 mol/L edetic acid, 300 mmol/L NaCl, 0.1% Igepal, 0.5 mmol/L NaF, 0.5 mmol/L Na3VO4, 0.5 mmol/L PMSF, and antiprotease mixture) using sonication. The lysates were centrifuged at 15 000×g for 30 min. Determination of protein concentrations of the supernatants was performed by the Bradford procedure (Bio-Rad Laboratory, Hercules, CA, USA). For STAT3 and pTry-stat3 analysis, the supernatant with 50 µg total protein was separated by electrophoresis on 10% SDS-Polyacrylamide gels and transferred onto PVDF membranes (Milipore, Bedford, MA) and blocked with 5% nonfat dry milk in PBS with 0.1% Tween-20. Blots were incubated with specific rabbit antibodies against STAT3 and pTry-stat3 and anti-β-actin antibody (Santa Cruz Biotech, Inc, Santa Cruz, CA, USA) and washed with TBST and subjected to corresponding HRP-conjugated secondary antibodies as indicated. For Bcl-2 analysis, 50 μg of total protein was electrophoresed on 12% SDS/PAGE gels and transferred onto PVDF membranes (Milipore, Bradford, MA,USA). The membranes were probed with mouse polyclonal antibodies against Bcl-2 antibody (Dako Biotech, Inc, Glostrup, Denmark) and washed with TBST and subjected to corresponding HRP-conjugated secondary antibodies as indicated. Blots were washed again with TBST and visualized by enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Uppsala, Sweden).
Proliferation and apoptosis assays in vitro Hep2 cells were incubated in 96-well plates. Cell proliferation was determined by 3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazo-lium bromide (MTT, Sigma) assay whereby cell numbers were counted by hemocytometer 72h after transfection. The absorbance values at 570 nm (A570) were determined on a multiwell plate reader. Cell growth inhibition rate was calculated according to the following formula:
A570c: A570 in control group; A570e: A570 in experimental group.
For FACS analysis of apoptosis, Hep2 cells were transfected with siRNA-STAT3 or pSilencer empty vector. After 72 h, cells were collected and washed with cold PBS containing edetic acid 4 mmol/L. Cells were fixed in 70% cold ethanol, collected by centrifugation, and washed once again with PBS containing edetic acid 4 mmol/L. Cells were resuspended in PBS containing edetic acid 4 mmol/L, 20 mL/L of propidium iodide (Sigma), 0.2% Triton X-100, 40 mg/L RNase A, and incubated for at least 30 min at 4 ºC. The cells were then analyzed by flow cytometry (FACScan, Becton Dickinson, Franklin Lakes, NJ, USA), using Cell Quest software. For fluoromicroscopic determination of positive apoptosis cells, 95 µL floating cells were mixed with 0.1% AO/EB [acridine orange (Sigma)/ethidium bromide (Sigma)] and observed under microscope.
Statistical analysis Data were expressed as mean±SD. The square χ2 analysis was performed to evaluate the significance of inter-group differences. Student’s t test was used for single comparison between two groups. Two-way ANOVA using the Student-Newman-Keuls method was adopted for comparison of variables after treatment. P<0.05 was considered significant. All statistical calculations were performed using SigmaStat statistical software package (SPSS10.0, Chicago, IL,USA).
Results
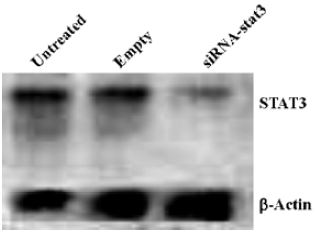
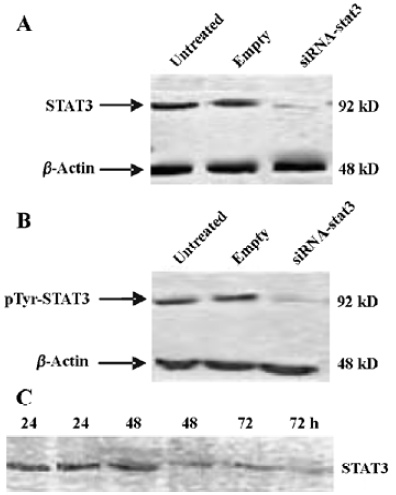
STAT3 RNAi by siRNA specifically reduces STAT3 expression in Hep2 cells Since STAT3 levels are significantly higher in tumor cells (including laryngeal tumor) than that in normal cells, we attempted to determine whether the synthetic STAT3 siRNA could inhibit the expression of STAT3 gene in Hep2 cells. Treatment of Hep2 cells with siRNA-STAT3 resulted in a significant decrease of STAT3 expression at both mRNA (Figure 1) and protein (Figure 2A) level compared to the untreated Hep2 cells and the vector-treated Hep2 cells, respectively. STAT3 expression was specifically targeted by STAT3 siRNA, since STAT3 siRNA unchanged the expression of β-actin (Figure 1, 2A). Of note is that pTyr-STAT3 was markedly expressed in untreated Hep2 cells and the vector-treated Hep2 cells (Figure 2B), indicating that the detected STAT3 was indeed in the form of tyrosine-phosphorylated(ie, activated) form. In the STAT3 siRNA Hep2 cells, Tyr-STAT3 expression was significantly reduced, indicating that STAT3 siRNA also inhibited the activity of STAT3. Furthermore, as shown by Figure 2C treatment of Hep2 cells with STAT3 siRNA caused time-dependent inhibitory effect on STAT3 expression in Hep2 cells.
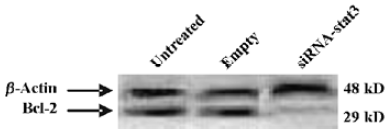
STAT3 siRNA down-regulates Bcl-2 expression in Hep2 cells Recent data indicate that constitutive activation of STAT3 induces the expression of anti-apoptotic genes including Bcl-2[17]. In order to determine whether Bcl-2 was involved in the STAT3-mediated apoptotic block in Hep2 cells, in this study Western blot analysis was performed. Figure 3 showed that Bcl-2 was remarkably expressed in the untreated Hep2 cells and the vector-treated Hep2 cells, whereas treatment of Hep2 cells with STAT3 siRNA significantly reduced the expression levels.
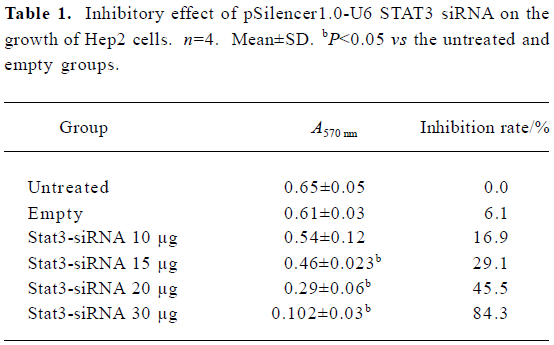
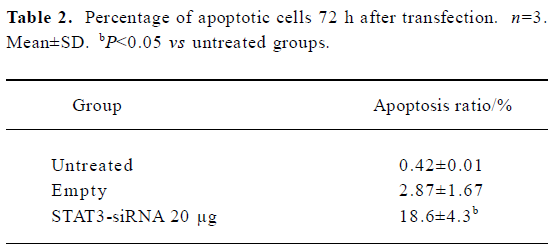
STAT3 siRNA inhibits growth and survival of Hep2 cells and induces apoptosis of Hep2 cells in vitro To determine whether synthetic STAT3 siRNA had an inhibitory effect on Hep2 cells growths, we accomplished determination of cell proliferation with MTT assay. Table 1 showed that treatment of Hep2 cells with STAT3 siRNA contributed to dose- dependent inhibition of Hep2 cells, whereas no inhibitory effect was observed in the untreated Hep2 cells and the vector-treated Hep2 cells.
Full table
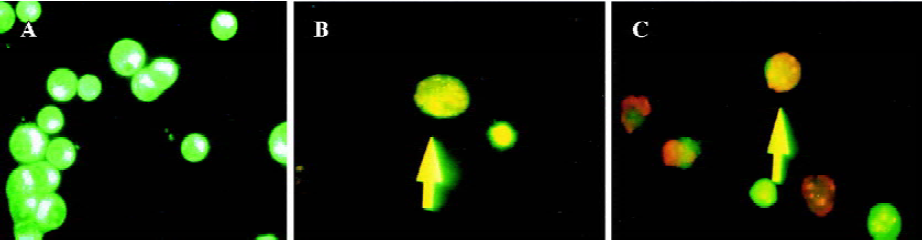
Two different methods including flow cytometry analysis and AO/EB staining (nucleus condensation) were adopted for determination of apoptosis in Hep2 cells (Table 2 and Figure 4). Flow cytometry analysis showed that in the STAT3 siRNA treated cells the apoptosis rate was significantly higher than that in the untreated Hep2 cells and the vector-treated Hep2 cells, whereas the apoptosis rate in the vector treated Hep2 was slightly higher than that in the untreated Hep2 cells but no significance was achieved (Table 2), indicating that siRNA-STAT3 induced apoptosis in Hep2 cells. Moreover, AO/EB staining revealed that both early apoptotic and late apoptotic Hep2 cells were seen in the cells treated with STAT3 siRNA, suggesting that STAT3 siRNA had not only an effect on induction of early apoptosis but also late apoptosis in Hep2 cells (Figure 3).
Full table
Discussion
Constitutively activated STAT3 is critical to STAT3 signaling pathway-dependent mechanism of malignancies[7,8,16]. A possible mechanism underlying transformation by activated STAT3 is the transcriptional upregulation of genes known to be involved in proliferation and apoptosis in laryngeal carcinomas and other malignant carcinomas. Under physiological conditions, STAT3 activation is transient and lasts from several minutes to several hours due to the transient nature of cytokine and growth factor signaling and the presence of proteins such as suppressor of cytokine signaling (SOCS) and PIAS (STAT blockade) that contribute to inhibition of STAT3 signaling. However, constitutive activation of protein tyrosine kinase (PTKs) occurs frequently during tumorigenesis due to activated mutations or aberrant growth factor or cytokine signaling, which results in constitutive activation of STAT3. Therefore, it is not surprising that constitutive activation of STAT3 is ubiquitous in human carcinomas[3,17,18]. In most instances, STAT3 that is capable of regulating growth, differentiation, and survival of cells is characterized by growth promoting manner[19–21]. At present, STAT3 has been recognized as an important onco-gene[2,21].
STAT3 is implicated in both embryogenesis and tummorigenesis. This may raise the question as to whether blocking STAT3 is beneficial for malignant carcinoma cells but harmful to normal cells. Takeda et al revealed that homozygous deletion of STAT3 was embryonically lethal[22]. Biochemical studies have shown that disruption of STAT3 signaling with dominant-negative approaches in murine fibroblasts does not inhibit normal cell growth[23,24]. Also, blockade of STAT3 signaling by decoy oligonucleotides in head and neck cancer cells only decreases the amount of STAT3 in normal cell but has no significant effect on cell viability.
Grandis et al[15] have reported that constitutive activation of STAT3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Treatment of tumor cells with inhibitors of STAT signaling results in decreased cell viability and induces apoptosis. Accumulated evidence has suggested that apoptotic regulatory proteins are implicated in STAT3 associated-apoptosis inhibition. It has been demonstrated that STAT3 regulates transcription from the Bcl-x promoter. Moreover, elevated expression levels of Bcl-xL mRNA in cells transformed by constitutively active STAT3 are observed[25].
Although the recently established approach of applying RNAi in mammalian studies by introducing siRNA can effectively inhibit gene expression[14,26,27], siRNA at present has been adopted as a promising genomic tool[17]. Such an approach overcomes many of the shortcomings previously experienced with approaches such as antibodies, antisense oligonucleotides, and pharmacological inhibitors. Intriguingly, RNAi targeting STAT3 by siRNA inhibits growth and induces apoptosis of prostate cancer and astrocytoma cells have been reported from different groups, however to date no one has known the effect of STAT3 siRNA on laryngeal cancer. Consequently, we have attempted to investigate the potential use of siRNA to block the expression of gene encoding STAT3 in laryngeal cancer.
To our knowledge we have successfully determined the inhibitory effect of the synthetic STAT3 siRNA on STAT3 expression in Hep2 cells at both mRNA and protein level (Figure 1, 2). We found that STAT3 was remarkably expressed in Hep2 cells, and treating Hep2 cells with STAT3 siRNA significantly reduced STAT3 expression levels characterized by a time-dependent inhibition of the gene expression. Increased STAT3 activation can occur through potential pathway of elevated constitutive levels of STAT3 protein and increased STAT3 tyrosine phosphorylation. In this study, the use of specific antibody of pTyr-STAT3 reveals that STAT3 siRNA inhibits not only STAT3 expression but also the activities of STAT3 in Hep2 cells (Figure 2), thereby confirming that the synthetic STAT3 siRNA can effectively inhibit STAT3 gene expressions in human laryngeal carcinoma cells. Our results are consistent with others who have shown the potent inhibitory effect of STAT3 siRNA on STAT3 expression in the models of prostate cancer and astrocytoma cell lines[16,17]. Moreover, other studies have shown that ubiquitous expression of STAT3 gene exhibits in a variety of human tumors and can be effectively inhibited by the introduction of STAT3 inhibitors, dominant negative STAT3, and/or blockade of tyrosine kinases besides STAT3 siRNA whereby treatment of tumors is achieved.
Recent data indicate that constitutive activation of STAT3 induces expression of anti-apoptotic genes including Bcl-2[17]. In the present study, to determine whether STAT3-mediated cell apoptosis in Hep2 cells exists, Western blot analysis was used for the measurement of Bcl-2 protein expression. Additionally, quantification of apoptotic cells by flow cytometry was also performed in this study. We found that apoptosis in Hep2 cells was arrested as evidenced by low percentages of apoptotic cells (0.42%±0.01% and 2.87%±1.67%) and remarkable expression levels of Bcl-2 protein, whereas treatment of Hep2 cells with STAT3 siRNA significantly increased the number of apoptotic cells (18.6%±4.3%, P<0.05) and decreased Bcl-2 protein expression levels (Table 2 and Figure 3). This suggests that STAT3 gene expression is an important implication in the regulation of apoptosis in Hep2 cells. Furthermore, visualization of nucleus condensation using AO/EB dyes suggests that the synthetic STAT3 siRNA has an effect on both early and late apoptosis. Our study is consistent with two recent intriguing reports where STAT3 siRNA was also adopted for the study of astrocytomas and human prostate cancer[16,17]. All are supportive for the proposed mechanism underlying STAT3 participating in oncogenesis is by inhibiting apoptosis through the induction of anti-apoptotic genes. Konnikova et al[17] have demonstrated that STAT3 is required for the expression of the anti-apoptotic genes survivin and Bcl-xL (a member of the Bcl-2 family of proteins) in astrocytoma cells. Likewise, Lee et al[16] have also shown that inhibition of STAT3 gene expression by siRNA induces apoptosis of human prostate cancer. Moreover, emerging evidence suggests that constitutive activation of STAT3 appears to be ubiquitous in tumors, which renders tumors cells resistant to apoptotic death caused by unbalanced expression level between anti-apoptotic genes and apoptotic genes[15,17,25]. Consistent with this, our recent work also has shown that human breast cancer cells implanted into nude mice exhibit remarkable expression of anti-apoptotic genes Bcl-2 but weak expression of apoptotic gene Bax accompanied by over-expression of STAT3 gene (Data not shown). The context is altered by treatment of the breast cancer with STAT3 siRNA characterized by unchanged expression of Bcl-2 genes, increased expression of apoptotic Bax gene, and significant inhibition of expression of STAT3 gene, and suppression of tumor growth (Data not shown).
siRNA has been effectively used in vivo to suppress gene expression in rats and adult mice whereby it achieves effective treatment of various organ and/or tissue disorders, including hepatitis, liver ischemia-reperfusion injury, allogeneic transplanted hepatocytes rejection, and CNS dis-orders[27-29]. This study also represents the first report that the synthetic STAT3 siRNA effectively suppresses Hep2 cells growths as evidenced by elevated inhibitory rate of Hep2 cell by STAT3 siRNA transfection (Table 1). In this study, an unexpected finding is that treatment of Hep2 cells with STAT3 siRNA is dose-dependent. Our results are consistent with other’s reports that STAT3 has the potential as a promising therapeutic molecular target in tumors including laryngeal cancer, astrocytomas, and human prostate cancer, thereby extrapolating that STAT3 siRNA may represent a novel approach in tumor gene therapy.
Strategies for producing siRNA duplexes include direct chemical synthesis, transcription with T7 promoter in vitro and recombinant DNA construction by vector with U6 promoter. Our results demonstrate that pSilencer1.0-U6 STAT3 siRNA can result in a long-term target-gene inhibition in Hep2 cells leading to growth suppression and induction of apoptosis in Hep2 cells. The STAT3 signaling pathway has been shown to be critical for the survival of a number of human tumors. This therefore raises the possibility that STAT3 siRNA could become an effective therapeutic agent for STAT3-dependent tumors.
Conclusion
This study represents the first report that demonstrates that STAT3 siRNA effectively inhibits STAT3 gene expression in Hep2 cells leading to growth suppression and induction of apoptosis in Hep2 cells. The use of siRNA technique may provide a novel therapeutic approach to treatment of laryngeal cancer and other malignant tumors expressing constitutively activated STAT3.
Acknowledgments
We thank Dr Bao-xue YANG from UCLA, CA, USA, for Northern blot analysis and Dr Yan MENG in the Department of Pathophysiology at the Basic School of Medicine of Jilin University for her assistance in siRNA design.
References
- Garcia R, Yu CL, Hudnall A, Catlett R, Nelson KL, Smithgall T, et al. Constitutive activation of STAT3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ 1997;8:1267-76.
- Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, et al. Constitutive activation of STAT3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 2001;20:2499-513.
- Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, et al. Requirement of STAT3 but not stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest 1998;102:1385-92.
- Takemoto S, Mulloy JC, Cereseto A, Migone TS, Patel BK, Matsuoka M, et al. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci USA 1997;94:13897-902.
- Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse JF, Capiod JC, Delobel J, et al. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood 1996;87:1692-7.
- Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi Sichuan, Mai JC, et al. Targeted inhibition of STAT3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA 2003;100:4138-43.
- Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, et al. Constitutive activation of STAT3 in human prostate tumors and cell lines: direct inhibition of STAT3 signaling induces apoptosis of prostate cancer cells. Cancer Res 2002;62:6659-66.
- Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of STAT3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 1999;10:105-15.
- Caldenhoven E, van Dijk TB, Solari R, Armstrong J, Raaijmakers JA, Lammers JW, et al. STAT3 beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem 1996;271:13221-7.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-strand RNA in Caenorhabditis elegans. Nature 1998;391:806-11.
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000;404:293-6.
- Sharp PA, Zamore PD. Molecular biology: RNA interference. Science 2000;287:2431-3.
- Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-strand RNA in early mouse development. Nat Cell Biol 2000;2:70-5.
- Elbashir S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494-8.
- Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, Endo S, et al. Constitutive activation of that stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci USA 2000;97:4227-32.
- Lee SO, Lou W, Qureshi KM, Mehraein-Ghomi F, Trump DL, Gao AC. RNA interference targeting Stat3 inhibits growth and induces apoptosis of human cancer cells. Prostate 2004;64:303-9.
- Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 2003;3:23.
- Weber-Nordt RM, Mertelsmann R, Finke J. The JAK-STAT pathway: signal transduction involved in proliferation, differentiation and transformation. Leuk Lymphoma 1998;28:459-67.
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of STAT3 in macrophages and neutrophils. Immunity 1999;10:39-49.
- Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA 1998;95:7556-61.
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. STAT3 as an oncogene. Cell 1999;98:295-303.
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, . Targeted disruption of the mouse STAT3 gene leads to early embryonic lethality. Proc Nat1 Acad Sci USA 1997; 94: 3801–4.
- Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, et al. Gene therapy with dominant-negative STAT3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res 1999;59:5059-63.
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute 2, a link between genetic and biochemical analyses of RNAi. Science 2001;293:1146-50.
- Scott S, Higdon R, Beckett L, Shi XB, deVere White RW, Earle JD, et al. Bcl 2 antisense reduces prostate cancer cell survival following irradiation. Cancer Biother Radiopharm 2002;17:647-56.
- Xia H, Mao QW, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol 2002;20:1006-10.
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Gen 2002;32:107-8.
- Song E, Lee SK, Wang J, Ince N, Ouyan N, Min J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003;9:347-51.
- Contreras JL, Vilatoba M, Eckstein C, Bilbao G, Thompson JA, Eckhoff DE. Caspase-8 and caspase-3 small interfering RNA decreases ischemia/reperfusion injury to the liver in mice. Surgery 2004;136:390-400.
