Development of a complex scintillation proximity assay for high-throughput screening of PPARγ modulators1
Introduction
Nuclear receptors (NRs) are a superfamily of ligand activated transcription factors that modulate specific gene expression. To date, more than 100 NRs have been identified including class I (ligand-dependent), class II (ligand-independent), and orphan receptors. A common feature of NRs is that they all contain a DNA binding domain that interacts with respective target genes to exert physiological functions[1]. Peroxisome proliferator-activated receptors (PPARs) with three isoforms (α, β, and γ) regulate gene transcription in response to small, lipophilic ligands[2-5]. PPARα is present in the liver, kidney, and heart, PPARβ (also known as PPARδ) is expressed ubiquitously, and PPARγ is mainly found in the adipose tissue and muscle. Upon ligand binding, PPARs release relevant co-repressors and form heterodimers with retinoid X receptors (RXRs)[6]. The heterodimers bind to peroxisome proliferator response elements (PPREs)[7,8] and recruit co-activators to initiate transcription of target genes. It is known that PPARγ is activated by fatty acids and prostaglandin J2 derivatives, although the identities of its physiologically relevant activators are not certain[9,10].
Because PPARγ activation can cause insulin sensitization, its synthetic agonists have been used in the treatment of type 2 diabetes[11,12]. Recently discovered liabilities of such therapy, namely, weight gain and edema, led to regulatory concerns on the long-term administration of drugs acting through PPARγ[13,14]. The elimination of such adverse effects may depend on the discovery of novel compounds with improved tissue selectivity while retaining insulin-sensiti-zing property.
Conventional methods to study and characterize NRs include non-homogeneous hydroxyapatite (HA) and gel shift assays which require a laborious separation procedure and thus, are not suitable for high-throughput screening (HTS). Scintillation proximity assay (SPA)[15] technology, howerer, provides a homogeneous screening approach that does not involve post-reaction liquid handling steps and is well-suited to automation and HTS. In the SPA system, an isotope (eg, [3H]) is brought very close to a scintillant-impregnated microbead or FlashPlate by binding to its surface. Because the emitted β particles or augur electrons can only travel short distances in the bulk solution, the microbead or FlashPlate preferentially captures electrons from the bound radiolabeled ligand. Therefore, the amount of light emitted from the scintillant in the microbead or FlashPlate is directly proportional to the amount of bound radiolabeled ligand (Figure 1). Several SPA-based NR competitive binding assays were developed and applied to HTS using biotinylated receptor ligand binding domains (LBDs; ‘ABC’ method)[16,17]. In this paper, we describe a more complex SPA-based assay system which includes the full-length PPARγ and RXRα, biotinylated PPRE, [3H]BRL49653 and streptavidin-coated FlashPlate or microbead in a homogeneous setting. This ‘ABCDE’ approach was fully validated and applied to HTS of a sizable compound library. A series of structurally diversified ‘hits’ were found, and subsequent characterization led to the discovery of two novel PPARγ binders with sub-micromolar potency and high specificity.
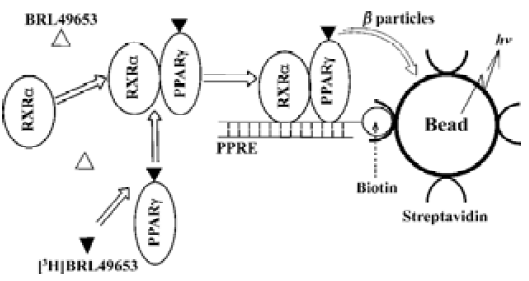
Materials and methods
Reagents Potassium chloride, sodium phosphate monobasic anhydrous, and magnesium chloride haxahydrate were purchased from Shanghai Chemical Co, Ltd. Edetic acid was purchased from Sigma-Aldrich (USA). BRL49653 and troglitazone were purchased from Cayman Chemical Co (USA). 3-[(3-Cholamidopropyl)dimethylammonio]-1-propane sulfonate (CHAPS) was purchased from Boehringer Mannheim GmbH (Germany). Dithiothreitol (DTT) was purchased from BioBasic Inc (Canada) and hydroxyapatite was obtained from Bio-Rad Laboratories (USA). Aprotinin and leupeptin were purchased from Merck KGaA (Germany). [3H]BRL49653 (53 Ci/mmol) was obtained from American Radiolabeled Chemicals, Inc (USA), FlashPlate and flat-bottom IsoplateTM was obtained from PerkinElmer, Inc (USA), and streptavidin-coated microbead was obtained from Amersham Biosciences UK Ltd (England). The plasmids of human NRs used in this study were from Dr Shen X of Shanghai Institute of Materia Medica, Chinese Academy of Sciences and Dr Chen SJ of Shanghai Institute of Hematology. Full-length PPARγ, RARα, β, γ, and RXRα, β, γ were produced with a baculovirus expression system using IPLB-Sf-21 cells[18]. The stock solutions for PPARγ, RARα, β, γ and RXRα, β, γ extract proteins were at 7, 12–15, and 7–13 g/L, respectively. Double-strand 5´-biotinylated-PPRE (CCTT-TGACCTATTGAACTATTACCT) was synthesized by Shanghai Sangon Biological Engineering Technology & Service Co, Ltd.
HA assay The assay buffer consists of 10% glycerol (v/v), NaH2PO4 25 mmol/L, MgCl2 0.5 mmol/L, DTT 1 mmol/L, edetic acid 1 mmol/L, CHAPS 5 mmol/L, aprotinin 2 mg/L and leupeptin 100 µmol/L. PPARγ 1 µL extract protein (70 mg/L) was loaded into each well of IsoplateTM containing the assay buffer, followed by [3H]BRL49653 (1.2 µL, 10 nmol/L) and various concentrations of BRL49653 or troglita-zone (2.5 µL), to give a final volume of 100 µL per well. The plates were sealed and incubated overnight at 4 oC. HA (25%, v/v) 25 mL was added to each well the next morning and the plates were gently agitated twice for 5 min each. Following centrifugation at 1200×g for 3 min, the supernatant was decanted and 100 µL assay buffer was added to each well. This washing procedure was repeated twice before adding 150 µL scintillation liquid (PerkinElmer), the plates were gently agitated to resuspend HA and counting was measured by a MicroBeta counter (PerkinElmer).
FlashPlate based SPA assay Biotinylated-PPRE (4 µL from a stock solution of 10 g/L) was mixed with the above assay buffer (20 mL) containing fish sperm DNA (Sangon; 20 µL from a 10 g/L stock solution), loaded to streptavidin-coated FlashPlate (200 µL/well) and incubated overnight at 4 oC. It was then washed three times with the assay buffer, 200 µL reaction solution containing 14 µg PPARγ extract protein (70 mg/L), 0.94 mg RXRα extract protein (4.7 mg/L), 10 nmol/L [3H]BRL49653 and various concentrations of BRL49653 or troglitazone were added to each well. Following incubation at 4 oC for 4 h, the plates were counted by the MicroBeta counter. For validation purpose, various concentrations of PPARγ and RXRα extract proteins, as well as different reaction time lengths, were studied to determine an optimal assay condition.
Microbead based SPA assay Biotinylated-PPRE (2 µL from a stock solution of 10 g/L) was mixed with the assay buffer (10 mL) containing fish sperm DNA (Sangon; 10 µL from a 10 g/L stock solution) and 4 mg streptavidin-coated microbead in a conical polypropylene centrifuge tube (Corning Inc, USA) and incubated overnight at 4 oC. The mixture was centrifuged for 10 min at 1500×g. The supernatant was then removed and washed three times with the 10-mL assay buffer. Reaction solution 10 mL containing 700 µg PPARγ extract protein (70 mg/L), 47 µg RXRα extract protein (4.7 mg/L), 10 nmol/L [3H]BRL49653 and various concentrations of BRL49653 or troglitazone were distributed to each well of a IsoplateTM (100 µL/well) and incubated at 4 oC for 4 h before counting by the MicroBeta counter. For validation purposes, various amounts of microbead were used to determine an optimal assay condi-tion.
HTS studies The compound library used for screening consists of 16 000 pure synthetic compounds and extracts of natural products. A 10-compound pool per well mix was applied to the primary screening (microbead based SPA assay), with an average concentration of 7 µmol/L for each compound dissolved in 100% Me2SO solution. This matrix system maximizes the advantage of HTS and allows duplicate screening of each compound[19]. In each 96-well IsoplateTM, 16 wells were used as positive control (BRL-49653) and samples showing greater than 70% inhibition were considered ‘hits’. Positive compounds were re-screened with FlashPlate based SPA assay and confirmed ‘hits’ studied for their binding cross-reactivities with RARα, β, γ and RXRα, β, γ using respective HA assay.
Results
Assay validation In the present study, we first assessed the kinetics of the signal strength generated by FlashPlate assay. Time-course experiment suggested that the equilibrium reached after 3.5 h of incubation at 4 oC and prolongation of the reaction time did not improve the assay efficiency (Figure 2A). Various concentrations of PPARγ and RXRα were used to establish the optimal assay condition. In the absence of unlabeled BRL49653, a maximum signal was detected with a combination of 140 mg/L (1:50 of the stock solution) PPARγ extract protein and 7 mg/L (1:1000 of the stock solution) RXRα (Figure 2B). The fully optimized assay possessed a signal to background ratio of 5. Thus, optimal receptor concentrations were determined for PPARγ (70 mg/L; 1:100) and RXRα (4.7 mg/L; 1:1500), respectively. Under this assay condition, IC50 values for the two PPARγ agonists, BRL49653 and troglitazone were measured (Figure 2C), and found to be comparable to those calculated from the HA assay (Figure 2D). Since the principle of microbead- based SPA assay is similar to that of FlashPlate, identical receptor concentrations were used with a reduced assay volume (100 µL/well). When different concentrations of microbead were used, a saturation reached between 2 and 4 g/L (Figure 3A) with a signal-to-background ratio equal to 5. The IC50 values for BRL49653 and troglitazone determined by this assay were within the range described earlier (Figure 3B).
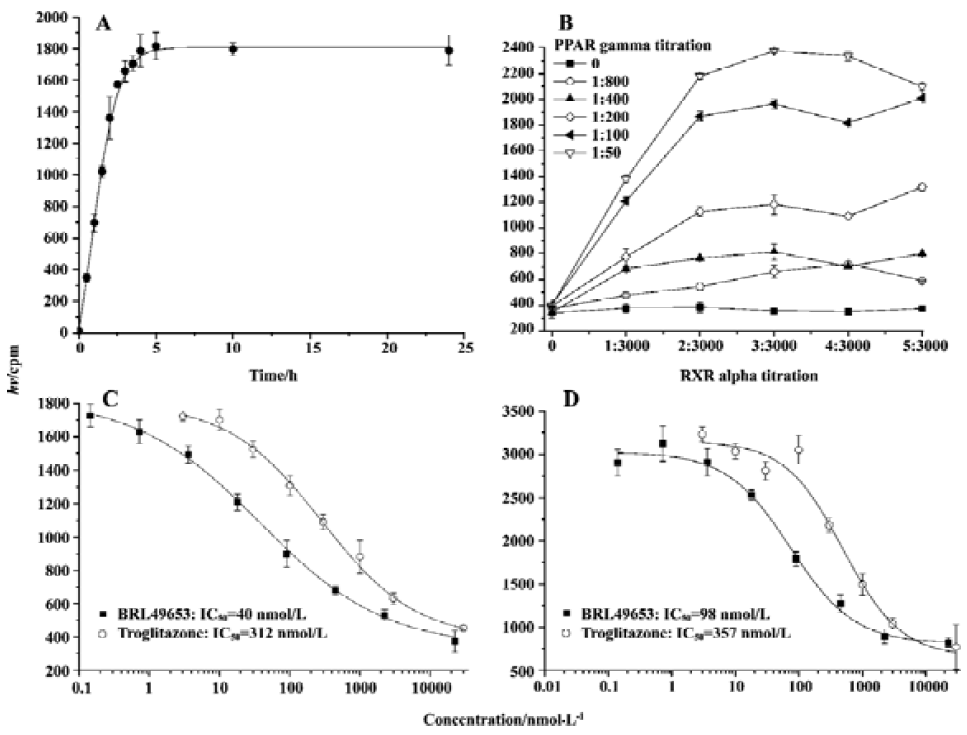
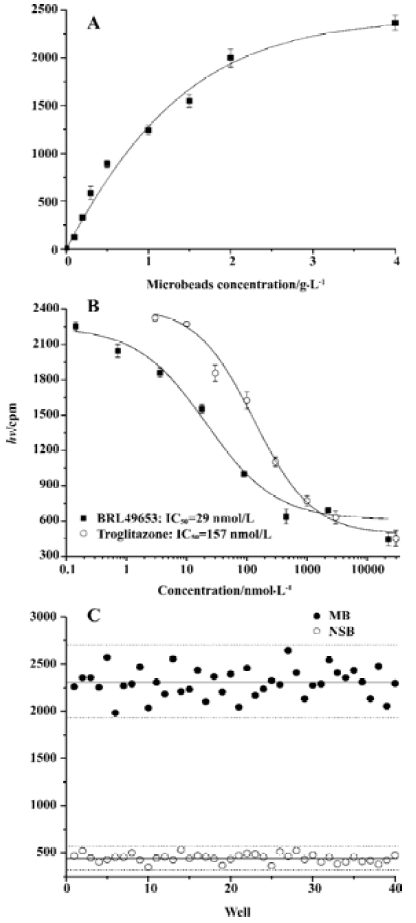
Assay parameters In order to apply the microbead-based SPA assay to HTS, both non-specific binding (NSB; using 22.5 mmol/L BRL49653) and maximum binding (MB; using 0 mmol/L BRL49653) were studied. Coefficient of variation (CV) values were 8.5% for NSB and 6.2% for MB, respectively (Figure 3C). The Z’ factor, which estimates the suitability to HTS[20], was calculated to be 0.71.
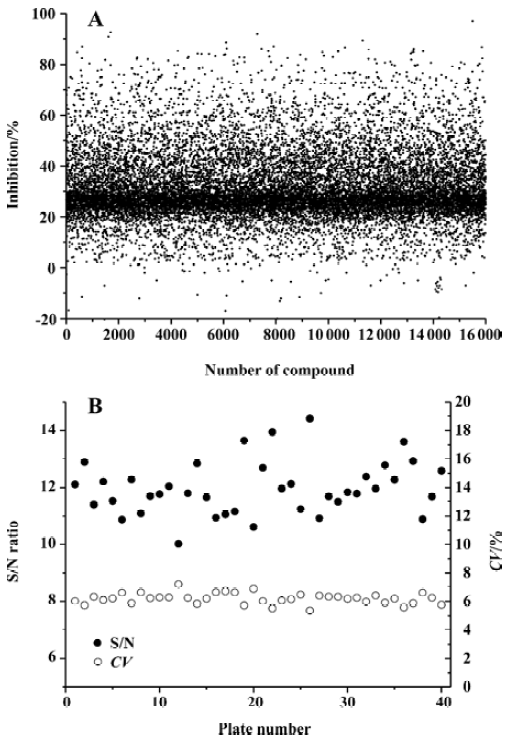
High-throughput screening Of the 16 000 samples initially screened, 178 ‘hits’ (1.11%) showing greater than 70% competitive inhibition on BRL49653 binding to PPARγ were discovered (all synthetic compounds; Figure 4A). Secondary (single compound per well) screening confirmed that 24 of the above ‘hits’ displayed consistent inhibitory effects with IC50 values between 0.2 and 28.5 μmol/L. Cross-reactivity studies with RARα, β, γ and RXRα, β, γ revealed that 12 of these compounds possess specific PPARγ binding properties including 2 with IC50 values less than 0.5 μmol/L (Table 1). In this HTS campaign, the signal-to-noise ratio (10-to-15-fold), CV (5%–8%) (Figure 4B) and Z’ factor (0.66–0.75) are of high quality nature.
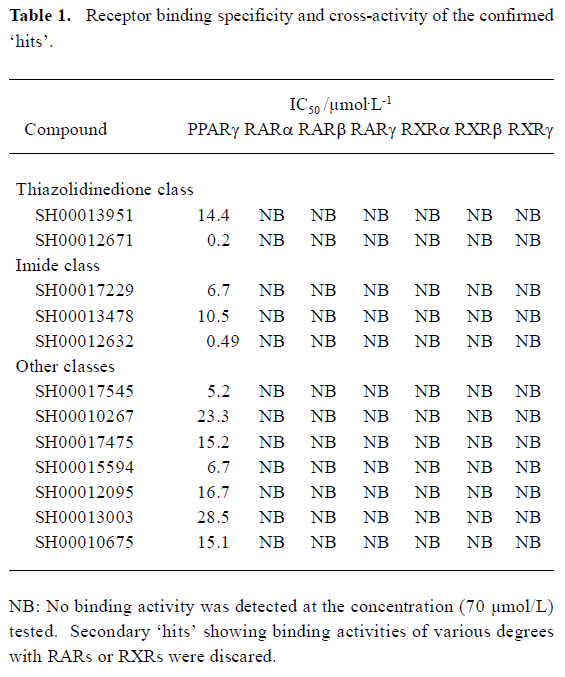
Full table
Discussion
Three receptor binding assays were employed and compared side-by-side in the present study to measure specific binding properties of two known PPARγ agonists, namely, BRL49653 and troglitazone. Conventional HA assay is a non-homogeneous method widely used to assess competitive interaction between a testing agent and receptor in the presence of radiolabeled ligand. For PPARγ, such interaction involves additional components such as RXRα and PPRE. Therefore, the ‘ABCDE’ method utilizing either FlashPlate or microbead based on SPA technology was developed and validated to include both RXRα and PPRE in the assay system. The final readout is fully dependent upon specific binding of biotinylated PPRE to streptavidin-coated FlashPlate or microbead as addition of free biotin was able to block this interaction completely[16]. Although the IC50 values of BRL49653 and troglitazone generated by these three approaches were very similar and comparable to those reported previously with a relatively simple ‘ABC’ SPA method (BRL49653: Kd=26 nmol/L, IC50=36 nmol/L; troglitazone: Kd=310 nmol/L, IC50=320 nmol/L)[16], the ‘ABCDE’ model is obviously superior in terms of physiologic mimicry, easy to use, robustness and efficiency, largely due to its inclusiveness and homogenous nature.
We found that both FlashPlate and microbead-based SPA methods could be readily adapted to automated HTS. However, the cost of FlashPlate, including assay volume (200 µL), associated reagent consumption, and compound depletion, etc, is approximately 5 times greater than that of microbead (100 µL). This may constitute a major concern when implementing a large HTS campaign. All the key assay parameters, such as CV and Z’ factor, obtained from our microbead-based SPA validation experiments, indicate that it is well suited to HTS[20]. Indeed, when employed in HTS of potential PPARγ modulators, this assay system demonstrated a consistently high quality in terms of the signal-to-noise ratio, CV and Z’ factor in all the 40 pooled compound matrix plates. One implication of this is that microbead-based SPA technology may be expanded to other NRs that form heterodimers upon activation.
Following the initial screening of 16 000 samples, a “hit” rate of 1.11% was achieved, of which, only 13.5% of the ‘hits’ could be confirmed by secondary screening. They were further tested for cross-reactivities with both RARs and RXRs (defined as IC50 less than 70 µmol/L), and 12 compounds showed high specificity for PPARγ with IC50 values ranging from 0.2 to 28.5 µmol/L. The IC50 for one thiazolidinedione-like compound is 0.2 µmol/L, better than several currently marketed PPARγ agonists. The other compound with an IC50 of 0.49 μmol/L belongs to the imide class. Both of them possess novel chemical structures (data not shown). If their activities could be demonstrated by cell-based functional assays, bioassay-guided structure modification and optimization may lead to the discovery of some entirely new PPARγ modulators.
Acknowledgement
We thank Dr Dale E MAIS, Xin XIE, Na LI, and Cheng-he JIN for their valuable discussions.
References
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell 1995;83:835-9.
- Schmidt A, Endo N, Rutledge SJ, Vogel R, Shinar D, Rodan GA. Identification of a new member of the steroid hormone receptor superfamily that is activated by a peroxisome proliferators and fatty acids. Mol Endocrinol 1992;6:1634-41.
- Sher T, Yi HF, McBride OW, Gonzalez FJ. cDNA cloning, chromosomal mapping and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry 1993;32:5598-604.
- Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev 1999;20:649-88.
- Wise H. Multiple signalling options for prostacyclin. Acta Pharmacol Sin 2003;24:625-30.
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 1992;358:771-4.
- Ijpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of PPAR-RXR heterodimer binding to DNA: a functional analysis of the malic enzyme gene PPRE. J Biol Chem 1997;272:20108-17.
- Juge-Aubry C, Pernin A, Favez T, Burger AG, Wahli W, Meier CA, et al. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements: importance of the 5’ flanking region. J Biol Chem 1997;272:25252-9.
- Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, et al. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem 1995;270:23975-83.
- Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehmann JM. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell 1995;83:813-9.
- O'Moore-Sullivan TM, Prins JB. Thiazolidinediones and type 2 diabetes: new drugs for an old disease. Med J Aust 2002;176:381-6.
- Yang L, An HX, Deng XL, Chen LL, Li ZY. Rosiglitazone reverses insulin secretion altered by chronic exposure to free fatty acid via IRS-2-associated phosphatidylinositol 3-kinase pathway. Acta Pharmacol Sin 2003;24:429-34.
- Camp HS. Thiazolidinediones in diabetes: current status and future outlook. Curr Opin Investig Drugs 2003;4:406-11.
- Guan Y, Zhang Y, Davis L, Breyer MD. Expression of peroxisome proliferator-activated receptors in urinary tract of rabbits and humans. Am J Physiol 1997;273:F1013-22.
- Hart HE, Greenwald EB. Scintillation proximity assay of antigen-antibody kinetics: concise communication. J Nucl Med 1979;20:1062-5.
- Nichols JS, Parks DJ, Consler TG, Blanchard SG. Development of a scintillation proximity assay for peroxisome proliferator-activated receptor gamma ligand binding domain. Anal Biochem 1998;257:112-9.
- Henke BR, Consler TG, Go N, Hale RL, Hohman DR, Jones SA, et al. A new series of estrogen receptor modulators that display selectivity for estrogen receptor beta. J Med Chem 2002;45:5492-505.
- Gearing KL, Göttlicher M, Teboul M, Widmark E, Gustafsson J. Interaction of the peroxisome proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci USA 1993;90:1440-4.
- Qian J, Voorbach MJ, Huth JR, Coen ML, Zhang HC, Ng SC, et al. Discovery of novel inhibitors of Bcl-xL using multiple high-throughput screening platforms. Anal Biochem 2004;328:131-8.
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high-throughput screening assays. J Biomol Screen 1999;4:67-73.
