Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique1
Introduction
Between the end of 2002 and June 2003, a severe epidemic disease called severe acute respiratory syndrome (SARS) broke out in China and quickly spread to more than 30 other countries. A novel coronavirus, SARS-CoV, was identified as the etiological agent of SARS infection by using biophysical and biochemical techniques[1–3]. Coronavirus (CoV) is a positive-stranded RNA virus and involves the largest viral RNA genome known to date. Phylogenetic studies have shown that SARS-CoV is a previously unknown coronavirus, which is neither a member nor a mutant of any known coronavirus group, and is believed to be a novel human coronavirus, possibly originating from a non-human host[4].
Proteolytic processing of viral polyproteins is a vital step in the replication cycle of many positive-strand RNA viruses and such processing is commonly performed by virus-genome encoded protease[5,6]. The open reading frame (ORF) of the coronavirus replicase gene for encoding the proteins which is required for virus replication and transcription, encompasses more than 20 000 nucleotides[7,8] and encodes two overlapping polyproteins, pp1a (replicase1a, around 450 kDa) and pp1ab (replicase1ab, approximately 750 kDa). It is known that the replicase gene features the sequence motifs of both papain-like protease and 3-chymotrypsin like protease (3CLpro)[9,10]. 3CLpro, which is also called main protease, functions as a key protease to control the activities of coronavirus replication complexes.
It has been concluded from previous research data that 3CLpro-mediated processing pathways are conserved in coronaviruses. Coronavirus main proteases employ cysteine and histidine residues as the catalytic dyad in the catalytic site but lack a corresponding third catalytic site[5,11–13], which is an acidic residue in chymotrypsin. Previous research has also confirmed that substrate specificities for the coronavirus main proteases are well defined, with the known proteolytic sites involving bulky hydrophobic residues (mainly leucine/isoleucine) at the P2 position, conserved glutamine at the P1 position, and small aliphatic residues at the P1’ posi-tion[14,15]. In addition, secondary structural studies for substrates of SARS-CoV 3CLpro have revealed that substrates with more beta-sheet like structures tend to be cleaved quickly[16]. The determination of the crystal structures for human coronavirus (strain 229E) 3CLpro and the inhibitor complex of porcine coronavirus (transmissible gastroenteritis virus, TGEV) 3CLpro also confirmed a remarkable degree of conservation of the substrate binding sites for coronavirus 3CLpro [17]. The recently reported crystal structures of SARS-CoV 3CLpro and its complex with an inhibitor revealed substantial pH-dependant conformational changes that correlate well with the varying activity of 3CLpro at different pH levels, and an unexpected model of inhibitor binding[18]. In fact, it has already been shown that 3CLpro is an ideal target for screening anti-virus agents[15,19,20]. Like other 3CLpro, SARS-CoV 3CLpro might become an attractive target in discovering new agents for the treatment of SARS[17].
In our previous work, we reported a 3D model of SARS-CoV 3CLpro with its inhibitors, designed by virtual screening[21], and the molecular cloning, expression and purification of SARS-CoV 3CLpro, with a preliminary study on its mass spectral characterization[22].
To date, the proteolytic activity of SARS-CoV 3CLpro has been almost determined by substrate-analog peptide cleavage assays using conventional RP-HPLC techniques[16]. In fact, the fluorescence-based assay is another method for quantitative protease activity assay, eg fluorescence resonance energy transfer (FRET) has been successfully used to develop spectrophotometric assays for many proteases[23–25]. The FRET-based method is more sensitive and less time-consuming compared with the RP-HPLC technique. Recently, a fluorogenic 14-amino acid peptide has been reported to measure SARS-CoV 3CLpro enzymatic activity[26]. In the present report, we describe how this methodology can be used to design a 12-amino acid fluorogenic peptide with EDANS/ Dabcyl as the fluorescence quenching pair. This fluorogenic substrate has been successfully used to characterize the proteolytic activities of wild type SARS-CoV 3CLpro at different pH levels and temperatures, and its four site-directed mutants including two catalytic residues and two substrate-binding sites as well. To our knowledge, such a fluorescence-based assay is the first to be used for site-directed mutation analysis of SARS-CoV 3CLpro. We hope that this present FRET-based assay will supply an ideal platform for the exploration of SARS-CoV 3CLpro putative inhibitors.
Materials and methods
Materials All chemicals were of HPLC grade and purchased from Sigma (St Louis, MO). The Ni-NTA chelating affinity column, protease for tag-cleavage and low molecular weight marker for SDS-PAGE were from Amersham Pharmacia Biotech (Uppsala, Sweden).
Cloning, expression and purification of SARS-CoV 3CLpro SARS-CoV 3CLpro was cloned, expressed, and purified, as described by Sun et al[22]. The purified His-tagged SARS-CoV 3CLpro was analyzed by SDS-PAGE, concentrated by centriprep (Milipore), and stored in sodium phosphate 20 mmol·L-1 pH 7.5/NaCl 100 mmol·L-1/dithiothreitol (DTT) 5 mmol·L-1/ethylene diaminetetraacetic acid (EDTA) 1 mmol·L-1 at 4 ºC. The structural integrity was analyzed by circular dichroism (CD).
Site-directed mutagenesis of SARS-CoV 3CLpro Site-directed mutagenesis was effected by using a modified recombinant PCR method. Four mutant SARS-CoV 3CLpro (SARS-CoV 3CLpro His41Ala, Cys145 Ala, His163Ala, and Met162Ala) were prepared with the QuickChange site-directed mutagenesis kit (Stratagene) using pQE30-SARS-CoV 3CLpro as a template. The nucleotide sequences of the primers used for site-directed mutagenesis were given in Table 1. The pQE30-SARS-CoV 3CLpro plasmids encoding mutant forms of SARS-CoV 3CLpro were verified by sequencing and then transformed into E coli M15 cells, and the mutant proteins were expressed and purified in a similar procedure to that for the wild type protease. The purity and structural integrity of the mutant proteins were analyzed by SDS-PAGE. The circular dichroism (CD) spectra of four site-directed mutants were compared with those of wild type SARS-CoV 3CLpro to exclude the possibility of structural misfolding caused by site-directed mutation (data not shown).
Full table
Synthesis of fluorogenic substrate The 12-amino acid fluorogenic substrate EDANS-Val-Asn-Ser-Thr-Leu-Gln-Ser-Gly-Leu-Arg-Lys(Dabcyl)-Met was synthesized and characterized using a modified procedure described by Garcia-Echeverria and Rich[27]. Nα-tert-butyloxycarbonyl (Boc)-protected amino acids were used in all coupling steps. The base liable-protecting group, 9-fluorenylmethyloxycar-bonyl (Fmoc), was used for the protection of the side chain of lysine, while the benzyl (Bzl) group was employed for the protection of C-terminal carboxyl group of the peptide. The Boc protecting group was removed before coupling by using HCl 4 mol/L in dioxane. Peptide couplings were achieved by overnight reaction with 1,3-dicyclohexylcarbodiimide (DCC) or 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride and 1-hydroxybenzotriazole (HOBt) in N,N-dimethylformamide (DMF). After completion of synthesis, the benzyl group was removed by hydrogenolysis in methanol. The commercially available 5-[(2´-aminoethyl)-amino] naphthelenesulfonic acid (EDANS) and 4-[[4-(dimethylamino) phenyl] azo] benzoic acid (Dabcyl) groups were attached to the peptides with conventional condensation reactions[28], and the Fmoc group was removed from the lysine ε-amine group by treatment with piperidine-N,N-dimethylformamide (1:1 v/v) to give the final fluorogenic substrate. The crude products were purified by means of HPLC on a Kromasil 7-μm C8 column (25 mm×250 mm). The purity of the final products was evaluated by reversed-phase HPLC on a Kromasil 5-μm C8 column (4.6 mm×250 mm). The integrity of the purified peptides was determined by LCQ-DECA mass spectrometry (Thermofinnigan, San Jose, CA), and the observed molecular mass was found to agree with the calculated value.
Enzymatic activity assay Stock solution for the fluoro-genic substrate was prepared in Me2SO and stored at 4 ºC. Subsequent dilutions were performed using the assay buffer (sodium phosphate 20 mmol/L, pH 7.5, NaCl 100 mmol/L, DTT 5 mmol/L, EDTA 1 mmol/L) with the final concentration of Me2SO less than 1% (v/v).
Initial fluorimetric assays were performed in a 1 mL quartz cuvette with a 1 cm path length at 25 ºC. During the assay, SARS-CoV 3CLpro (final concentration 1 μmol/L) was preincubated at 25 ºC for 30 min in the cuvette containing the assay buffer, followed by the addition of the fluorogenic substrate stock solutions to a final concentration of 10 μmol/L. The increase in emission fluorescence intensity was recorded at 10 min intervals on a Hitachi F-2500 fluorescence spectrophotometer connected to a thermostat. When fluorescence was being measured, the instrument was first set to zero with the fluorogenic substrate itself in the assay buffer. Cleavage of the peptide as a function of time was followed by monitoring the emission fluorescence intensity at a wavelength of 490 nm upon excitation at 340 nm (slit width 10 nm), and the initial reaction velocity (v0) was determined from the linear portion of the progress curve. The final emission fluorescence intensity of the totally hydrolyzed substrate was determined by adding excess SARS-CoV 3CLpro until no emission fluorescence intensity change at a wavelength of 490 nm was recorded.
Kinetic parameters (Km and kcat) of SARS-CoV 3CLpro for the fluorogenic substrate were determined by incubation of the substrate at different concentrations ranging from 1 mmol/L to 5 µmol/L with SARS-CoV 3CLpro 1 µmol/L at 25 ºC in the assay buffer. The reaction velocity (v0) for each substrate concentration was averaged from three assay results. Km and kcat values were calculated by using a Lineweaver-Burk plot.
The relative enzymatic activity at varying pH levels was investigated at 25 ºC in citric acid/phosphate buffer (pH =5, 6, 7, and 8) and glycine/NaOH buffer (pH=9, 10) containing DTT 5 mmol/L, EDTA 1 mmol/L, SARS-CoV 3CLpro 1 µmol/L and fluorogenic substrate 10 µmol/L. The reaction velocity (v0) at each pH value was measured in triplicate and averaged.
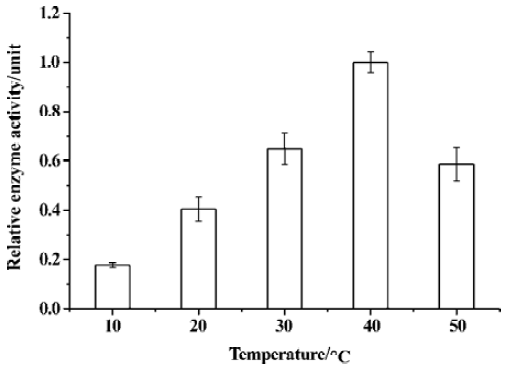
The enzymatic activity at different temperatures (10, 20, 30, 40, and 50 ºC) was measured in an assay buffer containing SARS-CoV 3CLpro 1 µmol·L-1and substrate 10 µmol/L. The enzyme was equilibrated at the same temperature as that for the assay buffer and substrate solution before mixing. During the assay, the cuvette temperature was stabilized by a thermostat connected to a Hitachi F-2500 fluorescence spectrophotometer. The reaction velocity (v0) at each temperature was the average of the three parallel assays and the reaction velocity at 40 ºC was taken as 1.0.
The enzymatic activity of four site-directed mutants of SARS-CoV 3CLpro (SARS-CoV 3CLpro His41Ala, Cys145 Ala, His163Ala, and Met162Ala) was the average of three parallel assays performed in an identical way as described above for wild type SARS-CoV 3CLpro.
Results and discussion
Fluorogenic substrate design and initial fluorimetric assay It is well estabished that the specificities of CoV 3CLpro for the substrate involves bulky hydrophobic residues (mainly leucine/isoleucine) at the P2 position, conserved glutamine at the P1 position, and small aliphatic residues at the P1’ position. A 12-amino acid peptide representing the NH2-terminal autoprocessing site of TGEV 3CLpro with the sequence of substrate-analog chloromethyl ketone inhibitor Cbz-Val-Asn-Ser-Thr-Leu-Gln-CMK was devised to separate the quencher from the fluorescent donor chromophore for designing the fluorogenic substrate in this study (Figure 1A), considering that a published cleavage experiment found that a 15-amino acid substrate involving these sequences could be efficiently cleaved by SARS-CoV 3CLpro[17]. To ensure efficient internal quenching, a commercially available donor/quencher pair was chosen for this study: 5-[(2´-aminoethyl)-amino] naphthelenesulfonic acid (EDANS) and 4-[[4-(dimethylamino) phenyl] azo] benzoic acid (Dabcyl) (Figure 1B). These dyes had already been used in FRET-based procedures[25,29,30]. Most importantly, the EDANS/Dabcyl pair possessed adequate spectral overlap which allowed almost complete quenching of EDANS’s fluorescence. The detailed experimental procedure for characterzing the enzymatic activity of SARS-CoV 3CLpro was illustrated in Figure 2.
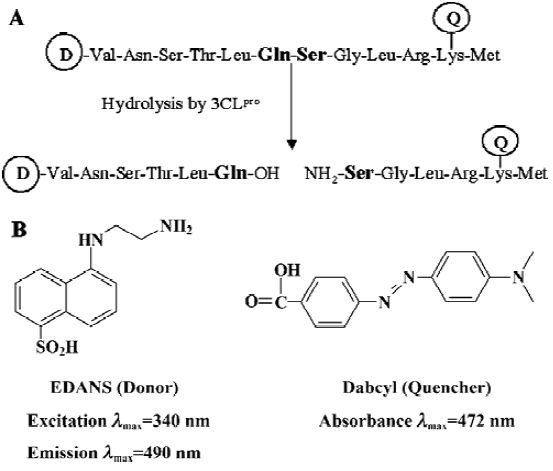
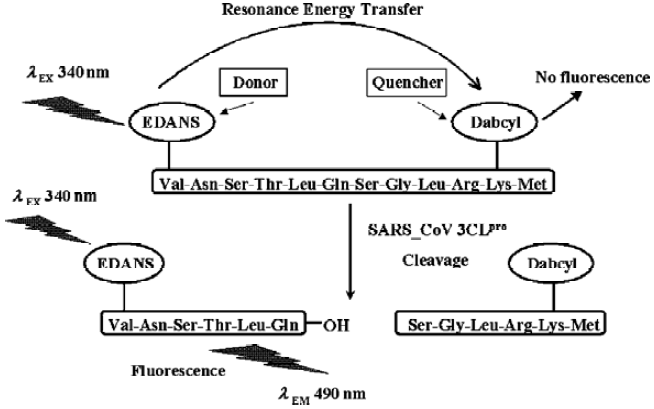
To evaluate the availability of the synthesized peptide as a potential substrate of SARS-CoV 3CLpro, an initial fluorimetric assay was performed. Comparison of the fluorescence emission spectrum of only the fluorogenic substrate (data not shown) with those spectra incubated with SARS-CoV 3CLpro (Figure 3A) clearly showed that the Dabcyl group almost exclusively quenched the donor emission of the EDANS fluorophore in the fluorogenic substrate and the fluorogenic substrate was efficiently hydrolyzed by SARS-CoV 3CLpro. The obvious emission fluorescence intensity enhancement over time implies that the fluorogenic substrate is ideal for subsequent enzymatic activity assays against SARS-CoV 3CLpro.
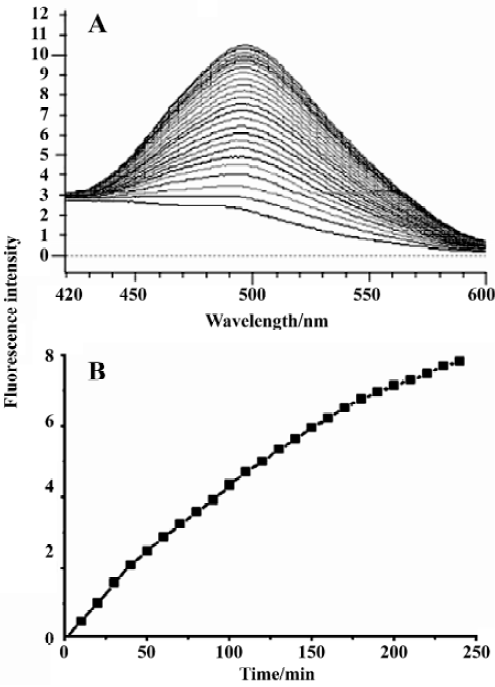
Enzymatic activity assays Considering the data obtained from the initial fluorimetric assay, SARS-CoV 3CLpro proteolysis against the fluorogenic substrate resulted in an appreciable increase in emission fluorescence intensity at a wavelength of 490 nm as a function of time, and a typical fluorescence profile following hydrolysis of the substrate is shown in Figure 3B. As a control, incubation of the substrate in assay buffer in the absence of SARS-CoV 3CLpro showed no fluorescence intensity change over time (data not shown).
Measurement of the kinetic parameters (Km and kcat) was accomplished by conducting hydrolysis of the fluorogenic substrate at various concentrations. A typical Lineweaver-Burk plot is shown in Figure 4, which plots the reciprocal of the initial velocity (v0) versus the reciprocal of the substrate concentration. The data for each concentration were obtained in triplicate. The calculated Km and kcat values were 404±9 μmol/L and 1.08±0.14 min-1, respectively. Obviously, the relatively small kcat/Km value (2.7±0.3 mmol-1·L·min-1) indicated that the in vitro activity of SARS-CoV 3CLpro was low, in common with other reported coronavirus 3CLpro[5]. Such a low activity for SARS-CoV 3CLpro may be due to the fact that only the dimer of 3CLpro was the active form, and efficient active dimeric form was often at relatively low concentrations during the enzyme assay[16,26].
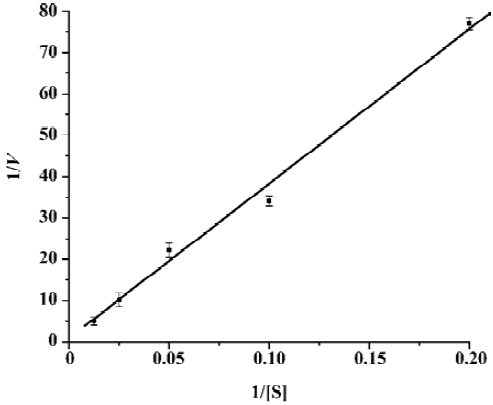
Figure 5 showed the results concerning the relative enzymatic activity at various pH values for SARS-CoV 3CLpro. The protease exhibited a stable proteolytic activity at pH 7.0–x9.0, and displayed only 50% activity at pH 6.0 and 10.0. However, when pH decreased to 5.0, it almost lost its activity completely. These results were strongly supported by a recent study, which discussed the pH dependence and the catalytic mechanism of SARS-CoV 3CLpro [31]. The pH profile shown in Figure 5 can be fitted by the following equation[32]:
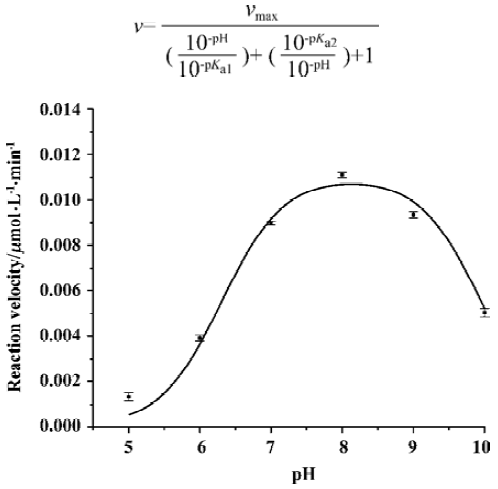
Where v is the measured reaction velocity that is plotted on the y axis, vmax is the observed maximum value of the reaction velocity, and pKa1 and pKa2 refer to the pKa values for the two relevant acid-base catalytic groups being titrated. A fit of the curve in Fig 5 to the equation yielded values of pKa1 and pKa2 of 6.31±0.12 and 9.95±0.11, respectivexxxand Cys145 formed a catalytic dyad in the active site of SARS-CoV 3CLpro, and ionization of sulfhydryl group of Cys145 and imidazole group of His41 played an important role in catalytic processing of substrate hydrolysis[18].
The relative enzymatic activity of SARS-CoV 3CLpro at different temperatures was depicted in Figure 6; the activity of SARS-CoV 3CLpro nearly doubled with every 10 ºC increase from 10 to 40 ºC, and such a result accorded well with the thermodynamics of a typical chemical reaction. Like all proteins, SARS-CoV 3CLpro undergoes thermal denaturation at elevated temperatures, hence the increases in the catalytic efficiency of the protease with increasing temperature might be compromised by the competing effects of the enzyme denaturation at high temperature. As indicated in Figure 6, the enzymatic activity diminished significantly at 50 ºC which was consistent with thermal denaturation of SARS-CoV 3CLpro as monitored by circular dichroism[16].
Mutation analysis of the catalytic dyad and two substrate-binding residues Based on the sequence alignments, previous mutagenesis studies with other CoV 3CLpro [7,9,33,34] and the recently reported crystal structure of SARS-CoV 3CLpro [18], the two residues His41 and Cys145 are fully conserved and considered to be the principal catalytic residues of CoV 3CLpro. Therefore, to confirm these two residues’ functions in the enzymatic activity of SARS-CoV 3CLpro, both residues were mutated into Ala. As indicated in Table 2, the proteolytic activity in SARS-CoV 3CLpro His41Ala and Cys145 Ala mutants were below the detection limit of the enzyme assay, which suggested a complete loss of enzymatic activities for these two mutants. Furthermore, these data strongly supported an indispensable catalytic function for His41 and Cys145, and were fully consistent with the mutagenesis data analysis published previously on the HCoV, IBV, MHV, and FIPV 3CLpro [5,11,35,36].
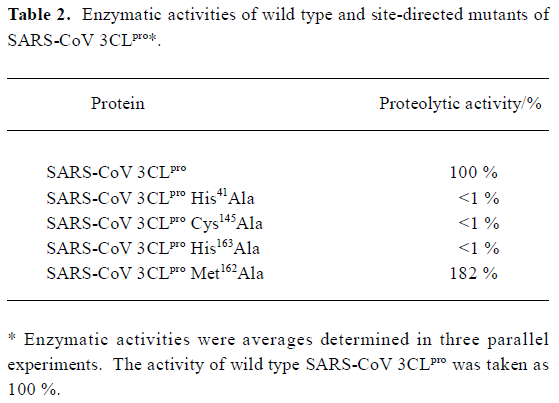
Full table
Additionally, it has been proposed that coronavirus 3CLpro might employ the characteristic sequence signature Tyr161-Met162-His163 for substrate binding[33]. However, to date, this theoretical functional character has not been tested experimentally for SARS-CoV 3CLpro. The need for experimental data becomes more evident considering the relatively low sequence identity between SARS-CoV 3CLpro and 3CLpro of other coronavirus groups. With these facts in mind, we focused on the Met162 and His163 residues of SARS-CoV 3CLpro by site-directed mutagenesis analysis.
First of all, we exchanged Ala for the conserved His163. As shown in Table 2, the proteolytic activity of this mutant dropped below the detection limit of the enzyme assay. Such an obvious indispensability of His163 for proteolytic activity supported the conclusion that His163 remained uncharged at physiological pH to facilitate optimal interaction with the Gln-P1 of the substrate in substrate-binding subsite S1 as determined by the recently reported crystal structure of SARS-CoV 3CLpro [18]. This was also consistent with the mutagenesis data obtained previously for the same His residue in other CoV 3CLpro[36]. Secondly, when Met162 residue was substituted with Ala, it was found that such a mutation did not decrease the proteolytic activity. On the contrary, it exhibited strongly increased activity: almost double compared with the wild type SARS-CoV 3CLpro. This result was consistent with the corresponding research for FIBV 3CLpro [36], and indicated that Met162 was not indispensable for proteolytic activity even though it was conserved among CoV 3CLpro. It was tentatively suggested that such an increase of proteolytic activity for SARS-CoV 3CLpro caused by Met162 mutation might be due to the fact that the substitution changed the spatial properties of the substrate-binding subsite S1 and facilitated substrate binding.
In summary, in this report, based on FRET theory, a fluorogenic substrate for detection of proteolytic activity of SARS-CoV 3CLpro was successfully synthesized and evaluated. The kinetic parameters showed that the in vitro activity of SARS 3CLpro was relatively low, perhaps due to the low concentration of the active dimeric form under the enzyme assay conditions. The pH profile of enzymatic activity yielded pKa1 and pKa2 values of 6.2 and 10.0, respectively, which correlated well with the finding that ionization of the catalytic dyad of His41 and Cys145 was perhaps vital during the catalytic processing of substrate hydrolysis. In addition, SARS-CoV 3CLpro displayed nearly a twofold increase in activity with every 10 ºC increase at 10–40 ºC, and diminished its activity significantly at 50 ºC owing to thermal denaturation. Mutation analysis revealed that substitution of either His41 and Cys145 residues resulted in complete loss of proteolytic activity, which was also observed for the residue His163 in the substrate-binding subsite S1. Replacement of Met162 with Ala caused increased activity, although the detailed mechanism was still unknown. The present FRET-based assay might supply an ideal platform for the exploration of SARS-CoV and other CoV 3CLpro putative inhibitors, given the conserved substrate specificities of CoV 3CLpro.
Footnote
Project supported by the State Key Program of Basic Research of China (grants 2003-CB514125, 2003CB514124, 2002CB512807, 2002CB512802, 2002AA233011), Sino-European Project on SARS Diagnostics and Antivirals (Proposal/Contract N
References
- Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest 2003;111:1605-9.
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319-25.
- Fouchier RA, Kuiken T, Schutten M, Van Amerongen G, Van Doornum GJ, Van Den Hoogen BG, et al. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 2003;423:240-7.
- Holmes KV. SARS-associated coronavirus. N Engl J Med 2003;348:1948-51.
- Ziebuhr J, Heusipp G, Siddell SG. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J Virol 1997;71:3992-7.
- Dougherty WG, Semler BL. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev 1993;57:781-822.
- Eleouet JF, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology 1995;206:817-22.
- Thiel V, Herold J, Schelle B, Siddell SG. Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol 2001;75:6676-81.
- Herold J, Raabe T, Schelle-Prinz B, Siddell SG. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology 1993;195:680-91.
- Lee HJ, Shieh CK, Gorbalenya AE, Koonin EV, Monica N, Tuler J, et al. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 1991;180:567-82.
- Liu DX, Brown TD. Characterization and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology 1995;209:420-7.
- Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in Nidovirales. J Gen Virol 2000;81:853-79.
- Hegyi A, Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J Gen Virol 2002;83:595-9.
- Ziebuhr J, Herold J, Siddell SG. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J Virol 1995;69:4331-8.
- Anand K, Plam GJ, Mesters JR, Siddell SG, Ziebuhr J, Higenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J 2002;21:3213-24.
- Fan KQ, Wei P, Feng Q, Chen SD, Huang CK, Ma L, et al. Biosyn-thesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem 2004;279:1637-42.
- Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 2003;300:1763-7.
- Yang HT, Yang MJ, Ding Y, Liu YW, Lou ZY, Zhou Z, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 2003;100:13190-5.
- Kim JC, Spence RA, Currier PF, Liu X, Denison MR. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine protease inhibitor E43d. Virology 1995;208:1-8.
- Someya Y, Takeda N, Miyamura T. Identification of active-site amino acid residues in the Chiba virus 3C-like protease. J Virol 2002;76:5949-58.
- Xiong B, Gui CS, Xu XY, Luo C, Chen J, Luo HB, et al. A 3D model of SARS-CoV 3CL proteinase and its inhibitors design by virtual screening. Acta Pharmacol Sin 2003;24:497-504.
- Sun HF, Luo HB, Yu CY, Sun T, Chen J, Peng SY, et al. Molecular cloning, expression, purification, and mass spectrometric characterization of 3C-like protease of SARS coronavirus. Protein Exp Purif 2003;32:302-8.
- Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metallopro-teinases. FEBS Lett 1992;296:263-6.
- Angliker H, Neumann U, Molloy SS, Thomas G. Internally quenched fluorogenic substrate for furin. Anal Biochem 1995;224:409-12.
- Mittoo S, Sundstrom LE, Bradley M. Synthesis and evaluation of fluorescent probes for the detection of calpain activity. Anal Biochem 2003;319:234-8.
- Kuo CJ, Chi YH, Hsu JT, Liang PH. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem Biophys Res Commun 2004;318:862-7.
- Garcia-Echeverria C, Rich DH. New intramolecularly quenched fluorogenic peptide substrates for the study of the kinetic specificity of papain. FEBS Lett 1992;297:100-2.
- Wang GT, Matayoshi E, Jan Huffaker H, Krafft GA. Design and synthesis of new fluorogenic HIV protease substrates based on resonance energy transfer. Tetrahedron Lett 1990;31:6493-6.
- Matayoshi ED, Wang GT, Krafft GA, Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 1990;247:954-8.
- Maggiora LL, Smith CW, Zhang ZY. A general method for the preparation of internally quenched fluorogenic protease substrates using solid-phase peptide synthesis. J Med Chem 1992;35:3727-30.
- Huang CK, Wei P, Fan KQ, Liu Y, Lai LL. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry 2004;43:4568-74.
- Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd ed. New York: Wiley-VCH Inc; 2000.
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res 1989;17:4847-61.
- Marra MA, Jones SJM, Astell CR, Holt RA, Wilson AB, Butterfield YSN, et al. The genome sequence of the SARS-associated coronavirus. Science 2003;300:1399-403.
- Lu Y, Denison MR. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology 1997;230:335-42.
- Hegyi A, Friebe A, Gorbalenya AE, Ziebuhr J. Mutational analysis of the active centre of coronavirus 3C-like proteases. J Gen Virol 2002;83:581-93.
