Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae1
Introduction
Three plasmid-mediated quinolone resistance genes, qnrA[1], qnrB[2], and qnrS[3], have been discovered since 1998. At least 6 qnrA, 6 qnrB, and 2 qnrS variants have been described[4]. qnrA was found in most common Enterobacteriaceae, including Escherichia coli (E coli), Klebsiella spp, Enterobacter spp, Citrobacter freundii, and Providencia stuartii [4], and it was located in complex sul1-type class 1 integrons[5]. qnrB and qnrS were identified in clinical strains of Klebsiella pneumoniae (K pneumoniae), E coli, Enterobacter spp, and Salmonella spp [4,6–9]. qnrB and qnrS were also detected in Citrobacter koseri[2]and Serratia marcescens[7], respectively.
Interestingly, qnrB and qnrS could be detected simultaneously in a few clinical strains[8,9]. Among 28 qnrA positive clinical strains of Enterobacteriaceae, 7 strains also harbored qnrS[10]. A further study was only carried out on a single strain of Enterobacter cloacae carrying 2 qnr genes simultaneously[9].
In China, qnrA has been detected in 8% of 78 clinical isolates of E coli[5], but qnrB and qnrS have not yet been reported in clinical strains. We found a clinical strain of K pneumoniae carrying both qnrB4 and qnrS1. The aim of this study was to identify the location and the relationship of the 2 genes, to analyze the genetic background of the qnrB4 and qnrS1 genes, and to evaluate their respective roles in the production of quinolone resistance.
Materials and methods
Bacterial strains A clinical strain of K pneumoniae carrying qnrB4 and qnrS1 was found during the study on extended-spectrum β-lactamases. The strain was isolated from sputum of an inpatient with acute exacerbation of chronic bronchitis at a teaching hospital in Shanghai in 2005. The patient was treated with cefradine, cefotaxime and levofloxacin prior to the isolation of K pneumoniae. Additional strains used were E coli V517[1]; E coli J53, containing plasmid R27[5] as standards for plasmid size; E coli J53AzR (resistant to azide)[5] as a recipient for conjugation; and E coli DH5α, which was used in cloning.
PCR detection The qnr genes (qnrA, qnrB, and qnrS), class A β-lactamase genes (blaTEM, blaSHV, blaPER, blaVEB, blaSFO, and blaCTX), and class C plasmid-mediated ampC β-lactamase genes were detected by PCR with specific primer sets in the clinical strain, transconjugants, and transformant. The primers used for qnrA, qnrB, and qnrS were 5'-GGG TAT GGA TAT TAT TGA TAA AG-3' and 5'-CTA ATC CGG CAG CAC TAT TA-3', 5'-ATG ACG CCA TTA CTG TAT AA-3' and 5'-GAT CGC AAT GTG TGA AGT TT-3', 5'-ACG ACA TTC GTC AAC TGC AA-3' and 5'-TAA ATT GGC ACC CTG TAG GC-3', respectively. The PCR conditions were 94°C for 45 s, 56°C for 45 s, and 72°C for 1min, and cycled 30 times for the detection of qnr genes. The primers used for CTX-M were 5'-AGT GCA AAC GGA TGA TGT-3' and 5'-GGC TGG GTA AAA ATA GGT C-3'. The primers for the ampC genes were previously described by Perez-Perez and Hanson[11] . All positive results were confirmed by direct sequencing of the PCR products on both strands.
Transfer of quinolone resistance Conjugation experiments were carried out in Luria-Bertani broth with azide-resistant E coli J53 as the recipient, as previously described[5] . Transconjugants were selected on Trypticase soy agar (TSA) plates containing sodium azide (200 mg/L) for counterselection and ampicillin (100 mg/L) or cefotaxime (8 mg/L) to select for plasmid-encoded resistance. Four hundred colonies were picked from the selection plates and detected by PCR for qnrB4 and qnrS1.
Transformation was performed for the qnrS1-bearing plasmid, which could not be transferred by conjugation. Plasmid DNA was extracted from the parent K pneumoniae strain using the QIAGEN plasmid midi kit (QIAGEN GmbH, Hilden, Germany) and introduced into electrocompetent E coli DH5α by electroporation. Colonies were selected on ciprofloxacin (0.06 mg/L). The colony carrying only 1 plasmid and harboring qnrS1 was confirmed with PCR. The qnrS1-bearing plasmid DNA was extracted again from the colony and introduced into E coli J53 by electroporation. Colonies were selected on plates containing 50 mg/L ampicillin, and were also confirmed carrying qnrS1 with PCR.
The plasmid size was estimated by agarose gel electro-phoresis, as previously described[5] ; the presence of qnrB4 and qnrS1 were confirmed with Southern blot hybridization using the DIG nucleic acid detection kit (Roche Applied Science, Mannheim, Germany).
In vitro susceptibility testing Minimal inhibitory concentrations (MIC) for the donor, recipient, transconjugant, and transformant strains were measured by agar dilution in accordance with the guidelines of the Clinical Laboratory Standards Institute (CLSI)[12] for ciprofloxacin, amikacin, ampicillin, cefepime, cefotaxime, ceftazidime, gentamicin, nalidixic acid, levofloxacin, sulfamethoxazole, and trimethoprim. The Etest (Biodisk AB, Solna, Sweden) was used to detect minimal changes in ciprofloxacin and levofloxacin susceptibility.
Cloning and nucleotide sequence analysis Plasmid DNA extracted from transconjugant or transformant strains harboring qnrB4 or qnrS1 were digested with HindIII, ligated to pUC18, and introduced into E coli DH5α with selection on TSA plates containing 50 mg/L ampicillin. Sequencing was carried out with an ABI Prism 3730 genetic analyzer (Applied Biosystems, Foster City, CA, USA), and was continued by primer walking on both DNA strands. For the sequence comparisons, the NCBI BLAST program (www.ncbi.nlm.nih.gov/blast/Blast.cgi) was utilized.
Nucleotide sequence accession numbers The nucleotide sequences in pHS7 and pHS8 containing qnrB4 and qnrS1, respectively, have been submitted to GenBank and have been assigned accession numbers EF683583 and EF683584, respectively.
Results
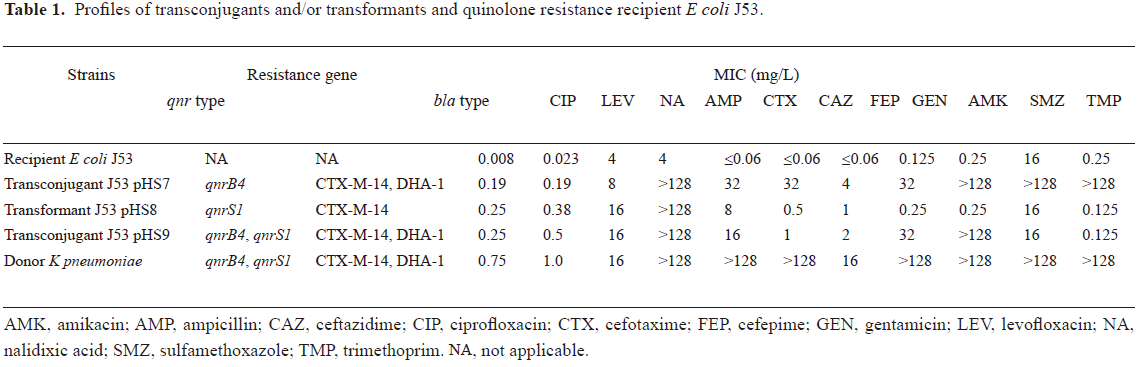
PCR detection and MIC determination of a clinical strain of K pneumoniae The clinical strain of K pneumoniae was identified as containing qnrB4, qnrS1, blaCTX-M-14, and blaDHA-1 genes by PCR and DNA sequencing. The clinical strain was susceptible to quinolones according to the CLSI criteria, with ciprofloxacin and levofloxacin MIC of 0.75 and 1.0 mg/L, respectively; but highly resistant to ampicillin, cefotaxime, ceftazidime, gentamicin, amikacin, sulfamethoxazole, and trimethoprim, and intermediate to cefepime (MIC 16 mg/L; Table 1).
Full table
Transfer of quinolone resistance and plasmid characterization qnrB4 and qnrS1 were located on 2 different plasmids, pHS7 and pHS8, and were 180 and 45 kb in size, respectively, by Southern blot hybridization. The qnrB4-bearing plasmid pHS7 could be transferred by conjugation to produce a transconjugant E coli J53 pHS7. Another transconjugant J53 pHS9 carrying plasmid pHS9 with a size similar to pHS7 bearing both qnrB4 and qnrS1, was also obtained by conjugation. qnrB4 was detected alone from 380 of 400 (95%) colonies picked from the selection plates by PCR; no qnrS1 alone was detected; qnrB4 and qnrS1 in combination were detected in 9 of 400 colonies (2.25%). The non-conjugative plasmid pHS8 bearing qnrS1 was transferred to J53 by transformation.
The ciprofloxacin MIC for J53 transconjugants or transformant carrying qnrB4 only (J53 pHS7), qnrS1 only (J53 pHS8), and both qnrB4 and qnrS1 (J53 pHS9) were 0.19, 0.25, and 0.25 mg/L, respectively. The MIC of levofloxacin for transconjugant J53 pHS8 and transformant J53 pHS9 were 0.38 and 0.5 mg/L, respectively, which were higher than that of J53 pHS7 (0.19 mg/L; Table 1).
Transconjugant J53 pHS7 was resistant to β-lactam antibiotics (ampicillin, cefotaxime, and ceftazidime), and pHS7 was found harboring blaCTX-M-14 and blaDHA-1 β-lactamase genes. Transformant J53 pHS8, also harboring the blaCTX-M-14 gene, was resistant to ampicillin and cefotaxime, but not to ceftazidime. Comparing the MICs of antimicrobials other than quinolones, we found that the resistance pattern in pHS9 was similar to that of pHS8, except that resistance to gentamicin and amikacin in pHS9, and resistance to sulfamethoxazole, trimethoprim, and ceftazidime in pHS7 was lost in pHS9 (Table 1).
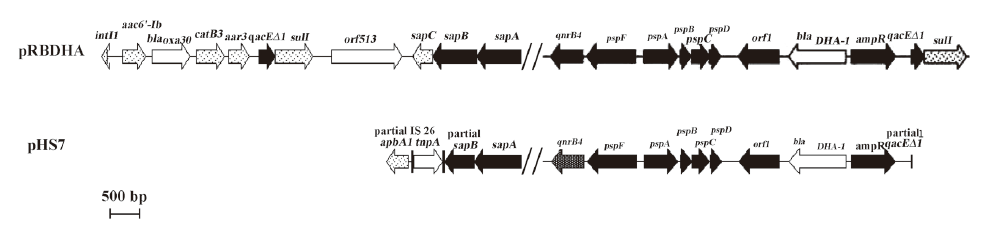
Analysis of plasmid structures The DNA sequences of plasmid pHS7 HindIII fragment showed that the upstream of qnrB4 included sapA and partial sapB, coding for putative peptide transport system permease, and aphA1. Notably, insertion sequence IS26 was located between aphA1 and sapB; downstream of qnrB4 included psp operons, coding for putative phage shock proteins, and ampC and ampR genes located between orf1 and partial qacEΔ1. The plasmid structure adjacent to qnrB4 was similar to that in plasmids, pRBDHA and pMPDHA[13], 2 qnrB4 and blaDHA-1-bearing plasmids (GenBank accession numbers AJ971343 and AJ971344, respectively), but found aphA1 and IS26 in the upstream of qnrB4 in pHS7 (Figure 1).
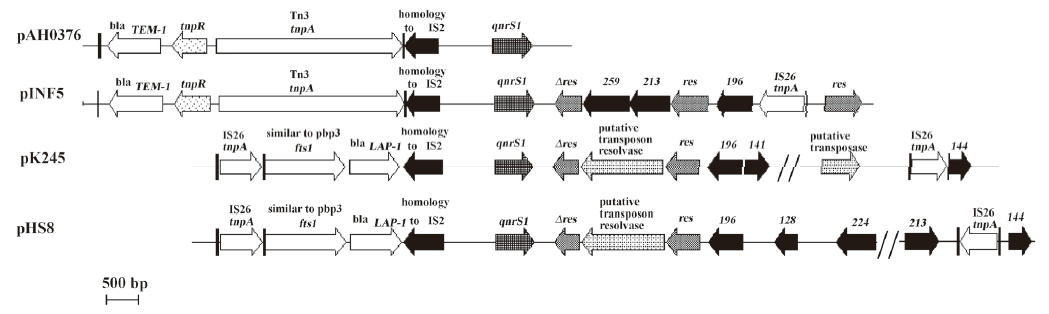
The sequence of the immediate region surrounding qnrS1 in pHS8 was nearly identical to the 3 reported qnrS1-bearing plasmids: pAH0376, pINF5, and pK245. Insertion sequence IS26 was found both upstream and downstream of qnrS1, and an IS2 was directly located upstream of qnrS1 (Figure 2).
Discussion
A clinical strain of K pneumoniae was identified as carrying both qnrB4 and qnrS1, and was isolated from a patient with acute exacerbation of chronic bronchitis. qnrB4 and qnrS1 were located on 2 separated plasmids, pHS7 and pHS8, respectively. pHS7 could be transferred to E coli J53 by conjugation. pHS8 could not be transferred alone by conjugation, but was successfully transferred to J53 by transformation. A recent study on 526 clinical strains isolated in Taiwan indicated that qnr genes were highly prevalent in E cloacae with a positive rate of 16% (86/526). Both qnrB2 and qnrS1 were detected simultaneously in 4 of the 86 qnr-positive strains, but there was no further study on the relationship, transferability, and location of the 2 qnr genes on these strains[8]. The only study was on an E cloacae clinical isolate co-expressing QnrB4 and QnrS1 determinants, isolated in France[9]. qnrB4 and qnrS1 were also located on 2 different plasmids, 100 and 160 kb in size, respectively, and could be transferred to E coli TOP10 by conjugation or transformation. The ciprofloxacin MIC of transconjugant or transformant carrying qnrB4 or qnrS1 were 0.06 and 0.12 mg/L, respectively. In our study, transformant J53 pHS8 carrying qnrS1 had higher MIC values for ciprofloxacin or levofloxacin (0.25–0.38 mg/L) than that of transconjugant J53 pHS7 carrying qnrB4 (0.19 mg/L). qnrS1 conferred higher quinolone MIC than qnrB4.
qnrA was located on complex sul1-type class 1 integrons, according to several reports[4,5,8]. qnrB was also located on sul1-type class 1 integrons from 2 reports[6,13]. A genetic environment analysis of qnrB2 in a Salmonella enterica Serovar Keurmassar showed that qnrB2 was located in a complex sul1-type integron which contained 2 class 1 integrons surrounding 2 common regions separated by a partial 3´ conserved segment[6]; the structure was similar to pMG252, the first qnrA-bearing plasmid[4]. During the study on the genetic organization of the ampC and ampR genes in Morganella morganii (M morganii), a qnrB4 gene was found to be located upstream of psp operons, and this was a complex sul1-type integron (qnrB4 was not labeled in the original figure, as it had not been reported at that time)[13]. In our study, the genetic environment of qnrB4 was similar to that of M morganii, but found aphA1 and IS26 upstream of qnrB4. Partial qacESΔ1 was found downstream of ampR, and intl1 gene and orf513 were detected by PCR (data not shown), so we supposed that qnrB4 was also located in a sul1-type class 1 integron in K pneumoniae.
Unlike qnrA and qnrB, qnrS was not located on any integrons according to 3 reported qnrS-bearing plasmid structure analyses[3,14,15], but qnrS was directly downstream of IS2. The PCR amplification was negative for intl1 and orf513 in qnrS-bearing plasmid pHS8 (data not shown), indicating that pHS8 did not carry any class 1 integron. Like in pK245, the fts1 and a blaLAP-1 genes were found upstream of qnrS1 in this study, and notably, insertion sequence IS26 was found both upstream and downstream of qnrS1. qnrS could also be located on a mobilizable incQ-related plasmid[16].
A transconjugant was obtained carrying plasmid pHS9, of a similar size to pHS7, but bore both qnrB4 and qnrS1. We supposed that there was an integration between pHS7 and pHS8 to produce pHS9 during the conjugation experiment. There was a possibility that a segment of plasmid pHS8 containing qnrS1 was integrated to pHS7. The blaLAP-1 gene that was located upstream of qnrS1 in pHS8, but not harbored by pHS7, was detected in pHS9 by PCR (data not shown). The movement of qnrS1 might be related to the insertion sequences of IS26 located upstream and downstream of qnrS1, and IS2 directly upstream of qnrS1. The resistance to sulfamethoxazole, trimethoprim, and ceftazidime in pHS7 was lost in pHS9, indicating that the qnrS1 segment might replace part of plasmid DNA in pHS7 or interrupt the expression of some genes in pHS7. However, the MIC of ciprofloxacin in pHS9 was not shown to be augmented from qnrB4 to qnrS1. Further study is ongoing on the analysis of the structure of pHS9 to understand the mechanism of the integration between the 2 plasmids.
This is the first report of qnrB and qnrS from China and these 2 genes are harbored by a single clinical strain of K pneumoniae.
Acknowledgements
We thank George A JACOBY and David C HOOPER for providing reference strains E coli J53AzR, E coli V517, and E coli R27. We are also grateful to George A JACOBY for critically reviewing the manuscript.
References
- Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet 1998;351:797-9.
- Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 2006;50:1178-82.
- Hata M, Suzuki M, Matsumoto M, Takahashi M, Sato K, Ibe S, et al. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother 2005;49:801-3.
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 2006;6:629-40.
- Wang M, Tran JH, Jacoby GA, Zhang YY, Wang F, Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother 2003;47:2242-8.
- Garnier F, Raked N, Gassama A, Denis F, Ploy MC. Genetic environment of quinolone resistance gene qnrB2 in a complex sul1-type integron in the newly described Salmonella enterica serovar Keurmassar. Antimicrob Agents Chemother 2006;50:3200-2.
- Poirel L, Leviandier C, Nordmann P. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother 2006;50:3992-7.
- Wu JJ, Ko WC, Tsai SH, Yan JJ. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother 2007;51:1223-7.
- Cattoir V, Poirel L, Nordmann P. Plasmid-mediated quinolone resistance determinant QnrB4 identified in France in an Enterobacter cloacae clinical isolate coexpressing a QnrS1 determinant. Antimicrob Agents Chemother 2007;51:2652-3.
- Lascols C, Robert J, Cattoir V, Bebear C, Cavallo JD, Podglajen I, et al. Type II topoisomerase mutations in clinical isolates of Enterobacter cloacae and other enterobacterial species harbouring the qnrA gene. Int J Antimicrob Agents 2007;29:402-9.
- Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbial 2002;40:2153-62.
- Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 15th informational supplement (M100-S15). Wayne, PA: Clinical Laboratory Standards Institute, 2005.
- Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother 2006;50:607-17.
- Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, et al. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 2006;50:3861-6.
- Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J Antimicrob Chemother 2006;58:18-22.
- Bonemann G, Stiens M, Puhler A, Schluter A. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob Agents Chemother 2006;50:3075-80.